Mismatch repair protein MutL becomes limiting during stationary-phase mutation
- PMID: 9308969
- PMCID: PMC316514
- DOI: 10.1101/gad.11.18.2426
Mismatch repair protein MutL becomes limiting during stationary-phase mutation
Abstract
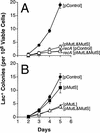
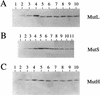
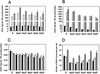
Postsynthesis mismatch repair is an important contributor to mutation avoidance and genomic stability in bacteria, yeast, and humans. Regulation of its activity would allow organisms to regulate their ability to evolve. That mismatch repair might be down-regulated in stationary-phase Escherichia coli was suggested by the sequence spectrum of some stationary-phase ("adaptive") mutations and by the observations that MutS and MutH levels decline during stationary phase. We report that overproduction of MutL inhibits mutation in stationary phase but not during growth. MutS overproduction has no such effect, and MutL overproduction does not prevent stationary-phase decline of either MutS or MutH. These results imply that MutS and MutH decline to levels appropriate for the decreased DNA synthesis in stationary phase, whereas functional MutL is limiting for mismatch repair specifically during stationary phase. Modulation of mutation rate and genetic stability in response to environmental or developmental cues, such as stationary phase and stress, could be important in evolution, development, microbial pathogenicity, and the origins of cancer.
Figures

References
-
- Anthoney DA, McIlwrath AJ, Gallagher WM, Edlin ARM, Brown R. Microsatellite instability, apoptosis, and loss of p53 function in drug-resistant tumor cells. Cancer Res. 1996;56:1374–1381. - PubMed
-
- Baker SM, Bronner CE, Zhang L, Plug AW, Robatzek M, Warren G, Elliott EA, Yu J, Ashley T, Arnheim N, Flavell RA, Liskay RM. Male mice defective in DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell. 1995;82:309–319. - PubMed
-
- Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie D-M, Monell C, Arnheim N, Bradley A, Ashley T, Liskay RM. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nature Genet. 1996;13:336–342. - PubMed
-
- Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW, Crosa JH, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. - PubMed
-
- Bremer H, Dennis PP. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt FC, Ingraham JL, Low KB, Magazanik B, Schaecter M, Umbarger E, editors. Escherichia coli and Salmonella typhimurium: Cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 1527–1542.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
