Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene
- PMID: 9314531
- PMCID: PMC2139824
- DOI: 10.1083/jcb.139.1.95
Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene
Abstract
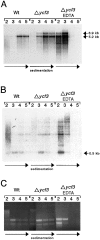
The chloroplast genome of all higher plants encodes, in its large single-copy region, a conserved open reading frame of unknown function (ycf3), which is split by two group II introns and undergoes RNA editing in monocotyledonous plants. To elucidate the function of ycf3 we have deleted the reading frame from the tobacco plastid genome by biolistic transformation. We show here that homoplasmic Deltaycf3 plants display a photosynthetically incompetent phenotype. Molecular analyses indicate that this phenotype is not due to a defect in any of the general functions of the plastid genetic apparatus. Instead, the mutant plants specifically lack detectable amounts of all photosystem I (PSI) subunits analyzed. In contrast, at least under low light conditions, photosystem II subunits are still present and assemble into a physiologically active complex. Faithful transcription of photosystem I genes as well as correct mRNA processing and efficient transcript loading with ribosomes in the Deltaycf3 plants suggest a posttranslational cause of the PSI-defective phenotype. We therefore propose that ycf3 encodes an essential protein for the assembly and/or stability of functional PSI units. This study provides a first example for the suitability of reverse genetics approaches to complete our picture of the coding capacity of higher plant chloroplast genomes.
Figures
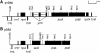




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

