CRP1, a LIM domain protein implicated in muscle differentiation, interacts with alpha-actinin
- PMID: 9314536
- PMCID: PMC2139825
- DOI: 10.1083/jcb.139.1.157
CRP1, a LIM domain protein implicated in muscle differentiation, interacts with alpha-actinin
Abstract
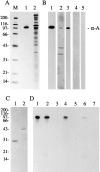
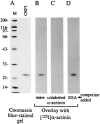
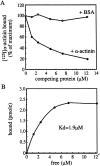
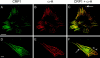
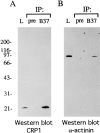
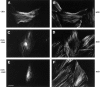
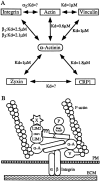
Members of the cysteine-rich protein (CRP) family are LIM domain proteins that have been implicated in muscle differentiation. One strategy for defining the mechanism by which CRPs potentiate myogenesis is to characterize the repertoire of CRP binding partners. In order to identify proteins that interact with CRP1, a prominent protein in fibroblasts and smooth muscle cells, we subjected an avian smooth muscle extract to affinity chromatography on a CRP1 column. A 100-kD protein bound to the CRP1 column and could be eluted with a high salt buffer; Western immunoblot analysis confirmed that the 100-kD protein is alpha-actinin. We have shown that the CRP1-alpha-actinin interaction is direct, specific, and saturable in both solution and solid-phase binding assays. The Kd for the CRP1-alpha-actinin interaction is 1.8 +/- 0.3 microM. The results of the in vitro protein binding studies are supported by double-label indirect immunofluorescence experiments that demonstrate a colocalization of CRP1 and alpha-actinin along the actin stress fibers of CEF and smooth muscle cells. Moreover, we have shown that alpha-actinin coimmunoprecipitates with CRP1 from a detergent extract of smooth muscle cells. By in vitro domain mapping studies, we have determined that CRP1 associates with the 27-kD actin-binding domain of alpha-actinin. In reciprocal mapping studies, we showed that alpha-actinin interacts with CRP1-LIM1, a deletion fragment that contains the NH2-terminal 107 amino acids (aa) of CRP1. To determine whether the alpha-actinin binding domain of CRP1 would localize to the actin cytoskeleton in living cells, expression constructs encoding epitope-tagged full-length CRP1, CRP1-LIM1(aa 1-107), or CRP1-LIM2 (aa 108-192) were microinjected into cells. By indirect immunofluorescence, we have determined that full-length CRP1 and CRP1-LIM1 localize along the actin stress fibers whereas CRP1-LIM2 fails to associate with the cytoskeleton. Collectively these data demonstrate that the NH2-terminal part of CRP1 that contains the alpha-actinin-binding site is sufficient to localize CRP1 to the actin cytoskeleton. The association of CRP1 with alpha-actinin may be critical for its role in muscle differentiation.
Figures





References
-
- Arber S, Caroni P. Specificity of single LIM motifs in targeting and LIM/LIM interactions in situ. Genes Dev. 1996;10:289–300. - PubMed
-
- Arber S, Halder G, Caroni P. Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell. 1994;79:221–231. - PubMed
-
- Arber S, Hunter JJ, Ross J, Hongo M, Sansig G, Borg J, Perriard J-C, Chien KR, Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88:393–403. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

