Initiation codon scanthrough versus termination codon readthrough demonstrates strong potential for major histocompatibility complex class I-restricted cryptic epitope expression
- PMID: 9314554
- PMCID: PMC2199058
- DOI: 10.1084/jem.186.7.1051
Initiation codon scanthrough versus termination codon readthrough demonstrates strong potential for major histocompatibility complex class I-restricted cryptic epitope expression
Abstract
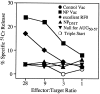
Accumulating evidence shows that the repertoire of major histocompatibility complex class I-restricted epitopes extends beyond conventional translation reading frames. Previously, we reported that scanthrough translation, where the initiating AUG of a primary open reading frame is bypassed, is most likely to account for the presentation of cryptic epitopes from alternative reading frames within the influenza A PR/8/34 nucleoprotein gene. Here, we confirm and extend these findings using an epitope cassette construct that features two well-defined CD8(+) T cell (TCD8+) epitopes in alternative reading frames, each preceded by a single start codon. Expression of one epitope depends on scanning of the ribosome over the first AUG with translation initiation occurring at the second AUG. We find that scanthrough translation has great potency in our system, with its impact being modulated, as predicted, by the base composition surrounding the first initiation codon, the number of start codons preceding the point of alternate reading frame initiation, and the efficiency with which the epitope itself is generated. Additionally, we investigated the efficiency of eukaryotic translation termination codons, to assess codon readthrough as a mechanism for cryptic epitope expression from 3' untranslated regions. In contrast with initiation codons, eukaryotic stop codons appear to be highly efficient at preventing expression of epitopes encoded in 3' untranslated regions, suggesting that 3' untranslated regions are not a common source of cryptic epitope substrate. We conclude that scanthrough is a powerful mechanism for the expression of epitopes encoded in upstream alternative open reading frames that may contribute significantly to TCD8+ responses and to tolerance induction.
Figures




References
-
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature (Lond) 1987;329:506–512. - PubMed
-
- Townsend A, Bodmer H. Antigen recognition by class I–restricted T lymphocytes. Annu Rev Immunol. 1989;7:601–624. - PubMed
-
- Yewdell JW, Bennink JR. Cell biology of antigen processing and presentation to major histocompatibility complex class I molecule-restricted T lymphocytes. Adv Immunol. 1992;52:1–123. - PubMed
-
- Heemels M-T, Ploegh H. Generation, translocation, and presentation of MHC class I–restricted peptides. Annu Rev Biochem. 1995;64:463–491. - PubMed
-
- York IA, Rock KL. Antigen processing and presentation by the class I major histocompatibility complex. Annu Rev Immunol. 1996;14:369–396. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

