Sphingomyelin depletion in cultured cells blocks proteolysis of sterol regulatory element binding proteins at site 1
- PMID: 9326582
- PMCID: PMC23408
- DOI: 10.1073/pnas.94.21.11179
Sphingomyelin depletion in cultured cells blocks proteolysis of sterol regulatory element binding proteins at site 1
Abstract
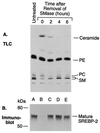
The current studies explore the mechanism by which the sphingomyelin content of mammalian cells regulates transcription of genes encoding enzymes of cholesterol synthesis. Previous studies by others have shown that depletion of sphingomyelin by treatment with neutral sphingomyelinase causes a fraction of cellular cholesterol to translocate from the plasma membrane to the endoplasmic reticulum where it expands a regulatory pool that leads to down-regulation of cholesterol synthesis and up-regulation of cholesterol esterification. Here we show that sphingomyelinase treatment of cultured Chinese hamster ovary cells prevents the nuclear entry of sterol regulatory element binding protein-2 (SREBP-2), a membrane-bound transcription factor required for transcription of several genes involved in the biosynthesis and uptake of cholesterol. Nuclear entry is blocked because sphingomyelinase treatment inhibits the proteolytic cleavage of SREBP-2 at site 1, thereby preventing release of the active NH2-terminal fragments from cell membranes. Sphingomyelinase treatment thus mimics the inhibitory effect on SREBP processing that occurs when exogenous sterols are added to cells. Sphingomyelinase treatment did not block site 1 proteolysis of SREBP-2 in 25-RA cells, a line of Chinese hamster ovary cells that is resistant to the suppressive effects of sterols, owing to an activating point mutation in the gene encoding SREBP cleavage-activating protein. In 25-RA cells, sphingomyelinase treatment also failed to down-regulate the mRNA for 3-hydroxy-3-methylglutaryl CoA synthase, a cholesterol biosynthetic enzyme whose transcription depends on the cleavage of SREBPs. Considered together with previous data, the current results indicate that cells regulate the balance between cholesterol and sphingomyelin content by regulating the proteolytic cleavage of SREBPs.
Figures



References
-
- Brown D A, Rose J K. Cell. 1992;68:533–544. - PubMed
-
- Schuchman E H, Desnick R J. In: The Metabolic and Molecular Basis of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors; Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw-Hill; 1995. pp. 2601–2624.
-
- Gatt S, Bierman E L. J Biol Chem. 1980;255:3371–3376. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

