A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity
- PMID: 9326604
- PMCID: PMC23448
- DOI: 10.1073/pnas.94.21.11301
A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity
Abstract
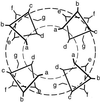
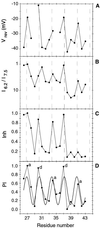
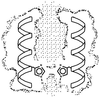
The M2 protein from influenza A virus forms proton-selective channels that are essential to viral function and are the target of the drug amantadine. Cys scanning was used to generate a series of mutants with successive substitutions in the transmembrane segment of the protein, and the mutants were expressed in Xenopus laevis oocytes. The effect of the mutations on reversal potential, ion currents, and amantadine resistance were measured. Fourier analysis revealed a periodicity consistent with a four-stranded coiled coil or helical bundle. A three-dimensional model of this structure suggests a possible mechanism for the proton selectivity of the M2 channel of influenza virus.
Figures


References
-
- Hille B. Ionic Channels of Excitable Membranes. Sunderland, MA: Sinauer; 1992.
-
- Song L, Hobaugh M R, Shustak C, Cheley S, Bayley H, Gouaux J E. Science. 1996;274:1859–1866. - PubMed
-
- Schulz G E. Curr Opin Struct Biol. 1996;6:485–490. - PubMed
-
- Unwin N. J Mol Biol. 1996;257:586–596. - PubMed
-
- Montal M. Curr Opin Struct Biol. 1996;6:499–510. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

