Functional association between promoter structure and transcript alternative splicing
- PMID: 9326631
- PMCID: PMC23504
- DOI: 10.1073/pnas.94.21.11456
Functional association between promoter structure and transcript alternative splicing
Abstract
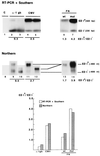
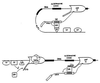
It has been assumed that constitutive and regulated splicing of RNA polymerase II transcripts depends exclusively on signals present in the RNA molecule. Here we show that changes in promoter structure strongly affect splice site selection. We investigated the splicing of the ED I exon, which encodes a facultative type III repeat of fibronectin, whose inclusion is regulated during development and in proliferative processes. We used an alternative splicing assay combined with promoter swapping to demonstrate that the extent of ED I splicing is dependent on the promoter structure from which the transcript originated and that this regulation is independent of the promoter strength. Thus, these results provide the first evidence for coupling between alternative splicing and promoter-specific transcription, which agrees with recent cytological and biochemical evidence of coordination between splicing and transcription.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

