Characterization of the CHD family of proteins
- PMID: 9326634
- PMCID: PMC23509
- DOI: 10.1073/pnas.94.21.11472
Characterization of the CHD family of proteins
Abstract
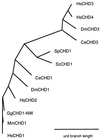
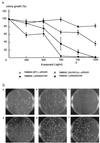
The murine gene CHD1 (MmCHD1) was previously isolated in a search for proteins that bound a DNA promoter element. The presence of chromo (chromatin organization modifier) domains and an SNF2-related helicase/ATPase domain led to speculation that this gene regulated chromatin structure or gene transcription. This study describes the cloning and characterization of three novel human genes related to MmCHD1. Examination of sequence databases produced several more related genes, most of which were not known to be similar to MmCHD1, yielding a total of 12 highly conserved CHD genes from organisms as diverse as yeast and mammals. The major region of sequence variation is in the C-terminal part of the protein, a region with DNA-binding activity in MmCHD1. Targeted deletion of ScCHD1, the sole Saccharomyces cerevesiae CHD gene, was performed with deletion strains being less sensitive than wild type to the cytotoxic effect of 6-azauracil. This finding suggested that enhanced transcriptional arrest at RNA polymerase II pause sites due to 6-azauracil-induced nucleotide pool depletion was reduced in the deletion strain and that ScCHD1 inhibited transcription. This observation, along with the known roles of other proteins with chromo or SNF2-related helicase/ATPase domains, suggests that alteration of gene expression by CHD genes might occur by modifications of chromatin structure, with altered access of the transcriptional apparatus to its chromosomal DNA template.
Figures



References
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

