Neurturin and glial cell line-derived neurotrophic factor receptor-beta (GDNFR-beta), novel proteins related to GDNF and GDNFR-alpha with specific cellular patterns of expression suggesting roles in the developing and adult nervous system and in peripheral organs
- PMID: 9334423
- PMCID: PMC6573771
- DOI: 10.1523/JNEUROSCI.17-21-08506.1997
Neurturin and glial cell line-derived neurotrophic factor receptor-beta (GDNFR-beta), novel proteins related to GDNF and GDNFR-alpha with specific cellular patterns of expression suggesting roles in the developing and adult nervous system and in peripheral organs
Abstract
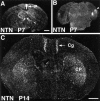
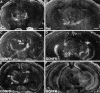
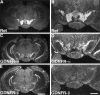
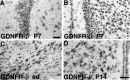
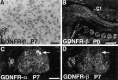
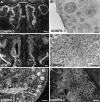
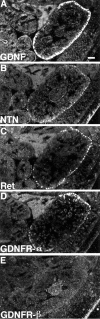
Cloning strategies were used to identify a gene termed glial cell line-derived neurotrophic factor receptor-beta (GDNFR-beta) related to GDNFR-alpha. In situ hybridization was then used to map cellular expression of the GDNF-related trophic factor neurturin (NTN) and GDNFR-beta mRNA in developing and adult mice, and comparisons with GDNFR-alpha and RET were made. Neurturin is expressed in postnatal cerebral cortex, striatum, several brainstem areas, and the pineal gland. GDNFR-beta mRNA was more widely expressed in the developing and adult CNS, including cerebral cortex, cerebellum, thalamus, zona incerta, hypothalamus, brainstem, and spinal cord, and in subpopulations of sensory neurons and developing peripheral nerves. NTN colocalized with RET and GDNFR-alpha in ureteric buds of the developing kidney. The circular muscle layer of the developing intestines, smooth muscle of the urether, and developing bronchiolae also expressed NTN. GDNFR-beta was found in myenteric but not submucosal intestinal plexuses. In developing salivary glands NTN had an epithelial expression, whereas GDNFR-beta was expressed in surrounding tissue. Neurturin and GDNFR-beta were present in developing sensory organs. In the gonads, NTN appeared to be expressed in Sertoli cells and in the epithelium of the oviduct, whereas GDNFR-beta was expressed by the germ cell line. Our findings suggest multiple roles for NTN and GDNFR-beta in the developing and adult organism. Although NTN and GDNFR-beta expression patterns are sometimes complementary, this is not always the case, suggesting multiple modi operandi of GDNF and NTN in relation to RET and the two binding proteins, GDNFR-alpha and GDNFR-beta.
Figures




References
-
- Angrist M, Bolk S, Halushka M, Lapchak P, Chakravarti A. Germline mutations in glial cell line-derived neurotrophic factor (GDNF) and RET in a Hirschsprung disease patient. Nat Genet. 1996;14:341–344. - PubMed
-
- Baloh R, Tansey M, Golden J, Creedon D, Heuckeroth R, Keck C, Zimonjic D, Popescu N, Johnson E, Jr, Milbrandt J. TrnR2, a novel receptor that mediates neurturin and GDNF signaling through Ret. Neuron. 1997;18:793–802. - PubMed
-
- Buj-Bello A, Adu J, Piñón L, Horton A, Thompson J, Rosenthal A, Chinchetru M, Buchman V, Davies A. Neurturin responsiveness requires a GPI-linked receptor and the Ret receptor tyrosine kinase. Nature. 1997;387:721–724. - PubMed
-
- Dagerlind Å, Friberg K, Bean A, Hökfelt T. Sensitive mRNA detection using unfixed tissue: combined radioactive and non-radioactive in situ hybridization histochemistry. Histochemistry. 1992;98:39–49. - PubMed
-
- Durbec P, Marcos-Gutierrez C, Kilkenny C, Grigoriou M, Suvanto P, Wartiovaara K, Smith D, Ponder B, Constantini F, Saarma M, Sariola H, Pachnis V. Glial cell line-derived neurotrophic factor signalling through the Ret receptor tyrosine kinase. Nature. 1996;381:789–792. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
