Prp43: An RNA helicase-like factor involved in spliceosome disassembly
- PMID: 9342317
- PMCID: PMC23592
- DOI: 10.1073/pnas.94.22.11798
Prp43: An RNA helicase-like factor involved in spliceosome disassembly
Abstract
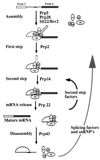
The Saccharomyces cerevisiae genes PRP2, PRP16, and PRP22 encode pre-mRNA splicing factors that belong to the highly conserved "DEAH" family of putative RNA helicases. We previously identified two additional members of this family, JA1 and JA2. To investigate its biological function, we cloned the JA1 gene and generated alleles carrying mutations identical to those found in highly conserved regions of other members of the DEAH family. A ja1 allele carrying a mutation identical to that in the temperature-sensitive (ts) prp22-1 gene conferred ts phenotype when integrated into the genome of a wild-type strain by gene replacement. Northern analysis of RNA obtained from the ts strain shifted to a nonpermissive temperature revealed accumulation of unspliced pre-mRNAs and excised intron lariats. Furthermore, analysis of splicing complexes showed that intron lariats accumulated in spliceosomes. The results presented indicate that JA1 encodes a pre-mRNA processing factor (Prp) involved in disassembly of spliceosomes after the release of mature mRNA. We have therefore renamed this gene PRP43.
Figures


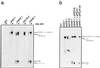

References
-
- Fuller-Pace F V, Lane D P. In: Nucleic Acids and Molecular Biology. Eckstein F, Lilley D M J, editors. Vol. 6. Berlin: Springer; 1992. pp. 159–173.
-
- Schmid S R, Linder P. Mol Microbiol. 1992;6:283–291. - PubMed
-
- Lamond A I. BioEssays. 1993;15:595–603. - PubMed
-
- Kramer A. In: Pre-mRNA Processing. Lamond I L, editor. Heidelberg: Springer; 1995. pp. 35–64.
-
- Madhani H D, Guthrie C. Annu Rev Genet. 1994;28:1–26. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

