Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation
- PMID: 9342321
- PMCID: PMC23611
- DOI: 10.1073/pnas.94.22.11819
Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation
Abstract
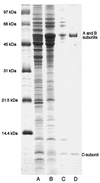
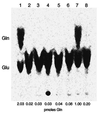
The three genes, gatC, gatA, and gatB, which constitute the transcriptional unit of the Bacillus subtilis glutamyl-tRNAGln amidotransferase have been cloned. Expression of this transcriptional unit results in the production of a heterotrimeric protein that has been purified to homogeneity. The enzyme furnishes a means for formation of correctly charged Gln-tRNAGln through the transamidation of misacylated Glu-tRNAGln, functionally replacing the lack of glutaminyl-tRNA synthetase activity in Gram-positive eubacteria, cyanobacteria, Archaea, and organelles. Disruption of this operon is lethal. This demonstrates that transamidation is the only pathway to Gln-tRNAGln in B. subtilis and that glutamyl-tRNAGln amidotransferase is a novel and essential component of the translational apparatus.
Figures





Comment in
-
Once there were twenty.Proc Natl Acad Sci U S A. 1997 Oct 28;94(22):11761-3. doi: 10.1073/pnas.94.22.11761. Proc Natl Acad Sci U S A. 1997. PMID: 9342308 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

