Targeting of NH2-terminal-processed microsomal protein to mitochondria: a novel pathway for the biogenesis of hepatic mitochondrial P450MT2
- PMID: 9348277
- PMCID: PMC2141697
- DOI: 10.1083/jcb.139.3.589
Targeting of NH2-terminal-processed microsomal protein to mitochondria: a novel pathway for the biogenesis of hepatic mitochondrial P450MT2
Abstract
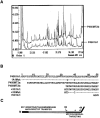
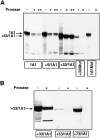
Cytochrome P4501A1 is a hepatic, microsomal membrane-bound enzyme that is highly induced by various xenobiotic agents. Two NH2-terminal truncated forms of this P450, termed P450MT2a and MT2b, are also found localized in mitochondria from beta-naphthoflavone-induced livers. In this paper, we demonstrate that P4501A1 has a chimeric NH2-terminal signal that facilitates the targeting of the protein to both the ER and mitochondria. The NH2-terminal 30-amino acid stretch of P4501A1 is thought to provide signals for ER membrane insertion and also stop transfer. The present study provides evidence that a sequence motif immediately COOH-terminal (residues 33-44) to the transmembrane domain functions as a mitochondrial targeting signal under both in vivo and in vitro conditions, and that the positively charged residues at positions 34 and 39 are critical for mitochondrial targeting. Results suggest that 25% of P4501A1 nascent chains, which escape ER membrane insertion, are processed by a liver cytosolic endoprotease. We postulate that the NH2-terminal proteolytic cleavage activates a cryptic mitochondrial targeting signal. Immunofluorescence microscopy showed that a portion of transiently expressed P4501A1 is colocalized with the mitochondrial-specific marker protein cytochrome oxidase subunit I. The mitochondrial-associated MT2a and MT2b are localized within the inner membrane compartment, as tested by resistance to limited proteolysis in both intact mitochondria and mitoplasts. Our results therefore describe a novel mechanism whereby proteins with chimeric signal sequence are targeted to the ER as well as to the mitochondria.
Figures










References
-
- Addya S, Zheng YM, Shayiq RM, Fan J, Avadhani NG. Characterization of a female-specific hepatic mitochondrial cytochrome P-450 whose steady-state level is modulated by testosterone. Biochemistry. 1991;30:8323–8330. - PubMed
-
- Anandatheerthavarada HK, Addya S, Dwivedi RS, Biswas G, Mullick J, Avadhani NG. Localization of multiple forms of inducible cytochromes P450 in rat liver mitochondria: immunological characteristics and patterns of xenobiotic substrate metabolism. Arch Biochem Biophys. 1997;339:136–150. - PubMed
-
- Andersson DJ, Mostov KE, Blobel G. Mechanisms of integration of de novo-synthesized polypeptides into membranes: signal recognition particle is required for integration into microsomal membranes of calcium ATPase of lens MP26 but not of cytochrome b5 . Proc Natl Acad Sci USA. 1983;80:7249–7253. - PMC - PubMed
-
- Andersson S, Davis DL, Dahlback H, Jomvall H, Russell DW. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

