Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form
- PMID: 9348288
- PMCID: PMC2141698
- DOI: 10.1083/jcb.139.3.717
Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form
Abstract
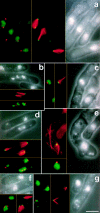
Through a screen designed to isolate novel fission yeast genes required for chromosome segregation, we have identified mal3+. The mal3-1 mutation decreased the transmission fidelity of a nonessential minichromosome and altered sensitivity to microtubule-destabilizing drugs. Sequence analysis revealed that the 35-kD Mal3 is a member of an evolutionary conserved protein family. Its human counterpart EB-1 was identified in an interaction screen with the tumour suppressor protein APC. EB-1 was able to substitute for the complete loss of the mal3+ gene product suggesting that the two proteins might have similar functions. Cells containing a mal3 null allele were viable but showed a variety of phenotypes, including impaired control of cell shape. A fusion protein of Mal3 with the Aequorea victoria green fluorescent protein led to in vivo visualization of both cytoplasmic and mitotic microtubule structures indicating association of Mal3 with microtubules. The absence of Mal3 protein led to abnormally short, often faint cytoplasmic microtubules as seen by indirect antitubulin immunofluorescence. While loss of the mal3+ gene product had no gross effect on mitotic spindle morphology, overexpression of mal3+ compromised spindle formation and function and led to severe growth inhibition and abnormal cell morphology. We propose that Mal3 plays a role in regulating the integrity of microtubules possibly by influencing their stability.
Figures







References
-
- Barbet N, Muriel WJ, Carr AM. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. . Gene (Amst) 1992;114:59–66. - PubMed
-
- Boguski MS, Lowe TMJ, Tolstoshev CM. dbEST-database for “Expressed Sequence Tags.” . Nat Genet. 1993;4:332–333. - PubMed
-
- Belmont LD, Mitchison TJ. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84:623–631. - PubMed
-
- Chang F, Wollard A, Nurse P. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile ring. J Cell Sci. 1996;109:132–142. - PubMed
-
- Cormack BP, Bertram G, Egerton M, Gow NA, Falkow S, Brown AJ. Yeast-enhanced green fluorescent protein (yEGFP) a reporter of gene expression in Candida albicans. Microbiology. 1997;143:303–311. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

