Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly
- PMID: 9348540
- PMCID: PMC25664
- DOI: 10.1091/mbc.8.10.2017
Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly
Abstract
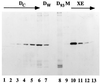
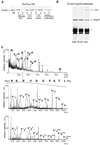
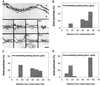
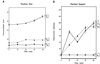
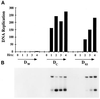
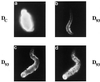
Yeast and vertebrate nuclear pores display significant morphological similarity by electron microscopy, but sequence similarity between the respective proteins has been more difficult to observe. Herein we have identified a vertebrate nucleoporin, Nup93, in both human and Xenopus that has proved to be an evolutionarily related homologue of the yeast nucleoporin Nic96p. Polyclonal antiserum to human Nup93 detects corresponding proteins in human, rat, and Xenopus cells. Immunofluorescence and immunoelectron microscopy localize vertebrate Nup93 at the nuclear basket and at or near the nuclear entry to the gated channel of the pore. Immunoprecipitation from both mammalian and Xenopus cell extracts indicates that a small fraction of Nup93 physically interacts with the nucleoporin p62, just as yeast Nic96p interacts with the yeast p62 homologue. However, a large fraction of vertebrate Nup93 is extracted from pores and is also present in Xenopus egg extracts in complex with a newly discovered 205-kDa protein. Mass spectrometric sequencing of the human 205-kDa protein reveals that this protein is encoded by an open reading frame, KIAAO225, present in the human database. The putative human nucleoporin of 205 kDa has related sequence homologues in Caenorhabditis elegans and Saccharomyces cerevisiae. The analyze the role of the Nup93 complex in the pore, nuclei were assembled that lack the Nup93 complex after immunodepletion of a Xenopus nuclear reconstitution extract. The Nup93-complex-depleted nuclei are clearly defective for correct nuclear pore assembly. From these experiments, we conclude that the vertebrate and yeast pore have significant homology in their functionally important cores and that, with the identification of Nup93 and the 205-kDa protein, we have extended the knowledge of the nearest-neighbor interactions of this core in both yeast and vertebrates.
Figures






Similar articles
-
Nup192p is a conserved nucleoporin with a preferential location at the inner site of the nuclear membrane.J Biol Chem. 1999 Aug 6;274(32):22646-51. doi: 10.1074/jbc.274.32.22646. J Biol Chem. 1999. PMID: 10428845
-
Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication.J Cell Biol. 1995 Mar;128(5):721-36. doi: 10.1083/jcb.128.5.721. J Cell Biol. 1995. PMID: 7876300 Free PMC article.
-
Identification of a new vertebrate nucleoporin, Nup188, with the use of a novel organelle trap assay.Mol Biol Cell. 2000 Oct;11(10):3381-96. doi: 10.1091/mbc.11.10.3381. Mol Biol Cell. 2000. PMID: 11029043 Free PMC article.
-
The diverse roles of the Nup93/Nic96 complex proteins - structural scaffolds of the nuclear pore complex with additional cellular functions.Biol Chem. 2014 May;395(5):515-28. doi: 10.1515/hsz-2013-0285. Biol Chem. 2014. PMID: 24572986 Review.
-
Nuclear pores and nuclear assembly.Curr Opin Cell Biol. 2001 Jun;13(3):363-75. doi: 10.1016/s0955-0674(00)00221-0. Curr Opin Cell Biol. 2001. PMID: 11343909 Review.
Cited by
-
Inhibition of Nuclear Pore Complex Formation Selectively Induces Cancer Cell Death.Cancer Discov. 2021 Jan;11(1):176-193. doi: 10.1158/2159-8290.CD-20-0581. Epub 2020 Sep 28. Cancer Discov. 2021. PMID: 32988961 Free PMC article.
-
The Ran GTPase cycle is required for yeast nuclear pore complex assembly.J Cell Biol. 2003 Mar 31;160(7):1041-53. doi: 10.1083/jcb.200209116. Epub 2003 Mar 24. J Cell Biol. 2003. PMID: 12654904 Free PMC article.
-
The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis.Mol Biol Cell. 2004 Jul;15(7):3333-44. doi: 10.1091/mbc.e03-12-0878. Epub 2004 May 14. Mol Biol Cell. 2004. PMID: 15146057 Free PMC article.
-
Nuclear envelope dysfunction and its contribution to the aging process.Aging Cell. 2020 May;19(5):e13143. doi: 10.1111/acel.13143. Epub 2020 Apr 15. Aging Cell. 2020. PMID: 32291910 Free PMC article. Review.
-
Clinical and Genetic Advances in Paget's Disease of Bone: a Review.Clin Rev Bone Miner Metab. 2017;15(1):37-48. doi: 10.1007/s12018-016-9226-0. Epub 2016 Dec 19. Clin Rev Bone Miner Metab. 2017. PMID: 28255281 Free PMC article. Review.
References
-
- Allende ML, Amsterdam A, Becker T, Kawakami K, Gaiano N, Hopkins N. Insertional mutagenesis in zebrafish identifies two novel genes, pescadillo and dead eye, essential for embryonic development. Genes Dev. 1996;10:3141–3155. - PubMed
-
- Becker J, Melchior F, Gerke V, Bischoff FR, Ponstingl H, Wittinghofer A. RNA1 encodes a GTPase-activating protein specific for Gsp1p, the Ran-TC4 homologue of Saccharomyces cerevisiae. J Biol Chem. 1995;270:11860–11865. - PubMed
-
- Blobel G, Potter VR. Nuclei from rat liver: isolation method that combines purity with high yield. Science. 1966;154:1662–1665. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

