RNA-peptide fusions for the in vitro selection of peptides and proteins
- PMID: 9356443
- PMCID: PMC24913
- DOI: 10.1073/pnas.94.23.12297
RNA-peptide fusions for the in vitro selection of peptides and proteins
Abstract
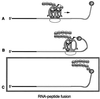
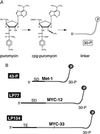
Covalent fusions between an mRNA and the peptide or protein that it encodes can be generated by in vitro translation of synthetic mRNAs that carry puromycin, a peptidyl acceptor antibiotic, at their 3' end. The stable linkage between the informational (nucleic acid) and functional (peptide) domains of the resulting joint molecules allows a specific mRNA to be enriched from a complex mixture of mRNAs based on the properties of its encoded peptide. Fusions between a synthetic mRNA and its encoded myc epitope peptide have been enriched from a pool of random sequence mRNA-peptide fusions by immunoprecipitation. Covalent RNA-peptide fusions should provide an additional route to the in vitro selection and directed evolution of proteins.
Figures



Comment in
-
mRNA display: diversity matters during in vitro selection.Proc Natl Acad Sci U S A. 2001 Apr 24;98(9):4825-6. doi: 10.1073/pnas.091101698. Proc Natl Acad Sci U S A. 2001. PMID: 11320229 Free PMC article. No abstract available.
References
-
- Breaker R R. Chem Rev (Washington, DC) 1997;97:371–390. - PubMed
-
- Osborne S E, Ellington A D. Chem Rev (Washington, DC) 1997;97:349–370. - PubMed
-
- Williams K P, Bartel D P. Nucleic Acids Mol Biol. 1996;10:367–381.
-
- Lorsch J R, Szostak J W. Acc Chem Res. 1996;29:103–110. - PubMed
-
- Gold L, Polisky B, Uhlenbeck O, Yarus M. Annu Rev Biochem. 1995;64:763–797. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

