The absence of effect of gid or mioC transcription on the initiation of chromosomal replication in Escherichia coli
- PMID: 9356478
- PMCID: PMC25015
- DOI: 10.1073/pnas.94.23.12497
The absence of effect of gid or mioC transcription on the initiation of chromosomal replication in Escherichia coli
Abstract
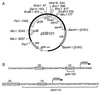
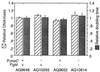
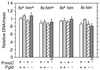
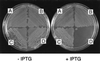
Despite the widely accepted view that transcription of gid and mioC is required for efficient initiation of cloned oriC, we show that these transcriptions have very little effect on initiation of chromosome replication at wild-type chromosomal oriC. Furthermore, neither gid nor mioC transcription is required in cells deficient in the histone-like proteins Fis or IHF. However, oriC that is sufficiently impaired for initiation by deletion of DnaA box R4 requires transcription of at least one of these genes. We conclude that transcription of mioC and especially gid is needed to activate oriC only under suboptimal conditions. We suggest that either the rifampicin-sensitive step of initiation is some other transcription occurring from promoter(s) within oriC, or the original inference of transcriptional activation derived from the rifampicin experiments is incorrect.
Figures
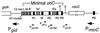
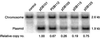
References
-
- Messer W, Weigel C. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1579–1601.
-
- Lark K G. J Mol Biol. 1972;64:47–60. - PubMed
-
- Tanaka M, Ohmori H, Hiraga S. Mol Gen Genet. 1983;192:51–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

