Association of INAD with NORPA is essential for controlled activation and deactivation of Drosophila phototransduction in vivo
- PMID: 9356510
- PMCID: PMC25084
- DOI: 10.1073/pnas.94.23.12682
Association of INAD with NORPA is essential for controlled activation and deactivation of Drosophila phototransduction in vivo
Abstract
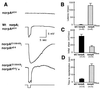
Visual transduction in Drosophila is a G protein-coupled phospholipase C-mediated process that leads to depolarization via activation of the transient receptor potential (TRP) calcium channel. Inactivation-no-afterpotential D (INAD) is an adaptor protein containing PDZ domains known to interact with TRP. Immunoprecipitation studies indicate that INAD also binds to eye-specific protein kinase C and the phospholipase C, no-receptor-potential A (NORPA). By overlay assay and site-directed mutagenesis we have defined the essential elements of the NORPA-INAD association and identified three critical residues in the C-terminal tail of NORPA that are required for the interaction. These residues, Phe-Cys-Ala, constitute a novel binding motif distinct from the sequences recognized by the PDZ domain in INAD. To evaluate the functional significance of the INAD-NORPA association in vivo, we generated transgenic flies expressing a modified NORPA, NORPAC1094S, that lacks the INAD interaction. The transgenic animals display a unique electroretinogram phenotype characterized by slow activation and prolonged deactivation. Double mutant analysis suggests a possible inaccessibility of eye-specific protein kinase C to NORPAC1094S, undermining the observed defective deactivation, and that delayed activation may similarly result from NORPAC1094S being unable to localize in close proximity to the TRP channel. We conclude that INAD acts as a scaffold protein that facilitates NORPA-TRP interactions required for gating of the TRP channel in photoreceptor cells.
Figures






Similar articles
-
Two distantly positioned PDZ domains mediate multivalent INAD-phospholipase C interactions essential for G protein-coupled signaling.EMBO J. 1998 Apr 15;17(8):2285-97. doi: 10.1093/emboj/17.8.2285. EMBO J. 1998. PMID: 9545241 Free PMC article.
-
The Calliphora rpa mutant lacks the PDZ domain-assembled INAD signalling complex.Eur J Neurosci. 2000 Nov;12(11):3909-18. doi: 10.1046/j.1460-9568.2000.00276.x. Eur J Neurosci. 2000. PMID: 11069586
-
The transient receptor potential protein (Trp), a putative store-operated Ca2+ channel essential for phosphoinositide-mediated photoreception, forms a signaling complex with NorpA, InaC and InaD.EMBO J. 1996 Dec 16;15(24):7036-45. EMBO J. 1996. PMID: 9003779 Free PMC article.
-
The roles of PDZ-containing proteins in PLC-beta-mediated signaling.Biochem Biophys Res Commun. 2001 Oct 19;288(1):1-7. doi: 10.1006/bbrc.2001.5710. Biochem Biophys Res Commun. 2001. PMID: 11594744 Review.
-
The organization of INAD-signaling complexes by a multivalent PDZ domain protein in Drosophila photoreceptor cells ensures sensitivity and speed of signaling.Cell Calcium. 1999 Nov;26(5):165-71. doi: 10.1054/ceca.1999.0070. Cell Calcium. 1999. PMID: 10643554 Review.
Cited by
-
Does Ca2+ reach millimolar concentrations after single photon absorption in Drosophila photoreceptor microvilli?Biophys J. 1999 Oct;77(4):1811-23. doi: 10.1016/S0006-3495(99)77026-8. Biophys J. 1999. PMID: 10512805 Free PMC article.
-
Light-regulated interaction of Dmoesin with TRP and TRPL channels is required for maintenance of photoreceptors.J Cell Biol. 2005 Oct 10;171(1):143-52. doi: 10.1083/jcb.200503014. J Cell Biol. 2005. PMID: 16216927 Free PMC article.
-
Vitamin A Deficiency Alters the Phototransduction Machinery and Distinct Non-Vision-Specific Pathways in the Drosophila Eye Proteome.Biomolecules. 2022 Aug 6;12(8):1083. doi: 10.3390/biom12081083. Biomolecules. 2022. PMID: 36008977 Free PMC article.
-
The TRP channel and phospholipase C-mediated signaling.Cell Mol Neurobiol. 2001 Dec;21(6):629-43. doi: 10.1023/a:1015191702536. Cell Mol Neurobiol. 2001. PMID: 12043838 Free PMC article. Review.
-
An unexpected INAD PDZ tandem-mediated plcβ binding in Drosophila photo receptors.Elife. 2018 Dec 10;7:e41848. doi: 10.7554/eLife.41848. Elife. 2018. PMID: 30526850 Free PMC article.
References
-
- Hurley J B. J Bioenerg Biomembr. 1992;24:219–226. - PubMed
-
- Selinger Z, Doza Y N, Minke B. Biochim Biophys Acta. 1993;1179:283–99. - PubMed
-
- Ranganathan R, Malicki D M, Zuker C S. Annu Rev Neurosci. 1995;18:283–317. - PubMed
-
- Lee Y-J, Dobbs M B, Verardi M L, Hyde D R. Neuron. 1990;5:889–898. - PubMed
-
- Lee Y-J, Shah S, Suzuki E, Zars T, O’Day P M, Hyde D R. Neuron. 1994;13:1143–1157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

