Mechanism of synergy between TATA and initiator: synergistic binding of TFIID following a putative TFIIA-induced isomerization
- PMID: 9367983
- PMCID: PMC316697
- DOI: 10.1101/gad.11.22.3007
Mechanism of synergy between TATA and initiator: synergistic binding of TFIID following a putative TFIIA-induced isomerization
Abstract
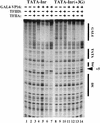
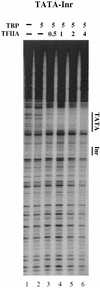
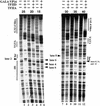
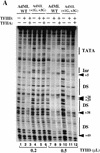
The TFIID complex interacts with at least three types of core promoter elements within protein-coding genes, including TATA, initiator (Inr), and downstream promoter elements. We have begun to explore the mechanism by which the TFIID-Inr interaction leads to functional synergy between TATA and Inr elements during both basal and activated transcription. In DNase I footprinting assays, GAL4-VP16 recruited TFIID-TFIIA to core promoters containing either a TATA box, an Inr, or both TATA and Inr elements, with synergistic interactions apparent on the TATA-Inr promoter. Appropriate spacing between the two elements was essential for the synergistic binding. Despite the sequence-specific TFIID-Inr interactions, gel shift experiments revealed that TFIID alone possesses similar affinities for the TATA-Inr and TATA promoters. Interestingly, however, recombinant TFIIA strongly and selectively enhanced TFIID binding to the TATA-Inr promoter, with little effect on binding to the TATA promoter. Studies of the natural adenovirus major late promoter confirmed these findings, despite the existence of specific but nonfunctional TFIID interactions downstream of the Inr in that promoter. These results suggest that a TFIIA-induced conformational change is essential for the sequence-specific TFIID-Inr interaction to occur with sufficient affinity to support the functional synergism between TATA and Inr elements.
Figures







References
-
- Burke TW, Kadonaga JT. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes & Dev. 1996;10:711–724. - PubMed
-
- Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. - PubMed
-
- Carey M, Leatherwood J, Ptashne M. A potent GAL4 derivative activates transcription at a distance in vitro. Science. 1990;247:710–712. - PubMed
-
- Chi T, Carey M. Assembly of the isomerized TFIIA–TFIID–TATA ternary complex is necessary and sufficient for gene activation. Genes & Dev. 1996;10:2540–2550. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
