u-shaped encodes a zinc finger protein that regulates the proneural genes achaete and scute during the formation of bristles in Drosophila
- PMID: 9367989
- PMCID: PMC316693
- DOI: 10.1101/gad.11.22.3083
u-shaped encodes a zinc finger protein that regulates the proneural genes achaete and scute during the formation of bristles in Drosophila
Abstract
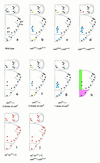
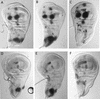
The pattern of the large sensory bristles on the notum of Drosophila arises as a consequence of the expression of the achaete and scute genes. The gene u-shaped encodes a novel zinc finger that acts as a transregulator of achaete and scute in the dorsal region of the notum. Viable hypomorphic u-shaped mutants display additional dorsocentral and scutellar bristles that result from overexpression of achaete and scute. In contrast, overexpression of u-shaped causes a loss of achaete-scute expression and consequently a loss of dorsal bristles. The effects on the dorsocentral bristles appear to be mediated through the enhancer sequences that regulate achaete and scute at this site. The effects of u-shaped mutants are similar to those of a class of dominant alleles of the gene pannier with which they display allele-specific interactions, suggesting that the products of both genes cooperate in the regulation of achaete and scute. A study of the sites at which the dorsocentral bristles arise in mosaic u-shaped nota, suggests that the levels of the u-shaped protein are crucial for the precise positioning of the precursors of these bristles.
Figures





References
-
- Bender W, Spierer P, Hogness DS. Chromosomal walking and jumping to isolate DNA from the Ace and rosy loci and the bithorax complex in Drosophila melanogaster. J Mol Biol. 1983;168:17–33. - PubMed
-
- Berg JM. Zinc finger proteins. Curr Opin Struct Biol. 1993;3:11–16.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
