Targeted deletion of alkylpurine-DNA-N-glycosylase in mice eliminates repair of 1,N6-ethenoadenine and hypoxanthine but not of 3,N4-ethenocytosine or 8-oxoguanine
- PMID: 9371767
- PMCID: PMC24230
- DOI: 10.1073/pnas.94.24.12869
Targeted deletion of alkylpurine-DNA-N-glycosylase in mice eliminates repair of 1,N6-ethenoadenine and hypoxanthine but not of 3,N4-ethenocytosine or 8-oxoguanine
Abstract
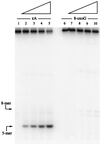
It has previously been reported that 1,N6-ethenoadenine (epsilonA), deaminated adenine (hypoxanthine, Hx), and 7,8-dihydro-8-oxoguanine (8-oxoG), but not 3,N4-ethenocytosine (epsilonC), are released from DNA in vitro by the DNA repair enzyme alkylpurine-DNA-N-glycosylase (APNG). To assess the potential contribution of APNG to the repair of each of these mutagenic lesions in vivo, we have used cell-free extracts of tissues from APNG-null mutant mice and wild-type controls. The ability of these extracts to cleave defined oligomers containing a single modified base was determined. The results showed that both testes and liver cells of these knockout mice completely lacked activity toward oligonucleotides containing epsilonA and Hx, but retained wild-type levels of activity for epsilonC and 8-oxoG. These findings indicate that (i) the previously identified epsilonA-DNA glycosylase and Hx-DNA glycosylase activities are functions of APNG; (ii) the two structurally closely related mutagenic adducts epsilonA and epsilonC are repaired by separate gene products; and (iii) APNG does not contribute detectably to the repair of 8-oxoG.
Figures

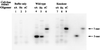



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

