Cloning and expression of rat 25-hydroxyvitamin D3-1alpha-hydroxylase cDNA
- PMID: 9371776
- PMCID: PMC24239
- DOI: 10.1073/pnas.94.24.12920
Cloning and expression of rat 25-hydroxyvitamin D3-1alpha-hydroxylase cDNA
Abstract
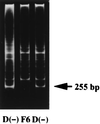
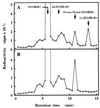
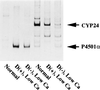
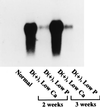
A full-length cDNA for the rat kidney mitochondrial cytochrome P450 mixed function oxidase, 25-hydroxyvitamin D3-1alpha-hydroxylase (P4501alpha), was cloned from a vitamin D-deficient rat kidney cDNA library and subcloned into the mammalian expression vector pcDNA 3.1(+). When P4501alpha cDNA was transfected into COS-7 transformed monkey kidney cells, they expressed 25-hydroxyvitamin D3-1alpha-hydroxylase activity. The sequence analysis showed that P4501alpha was of 2,469 bp long and contained an ORF encoding 501 amino acids. The deduced amino acid sequence showed a 53% similarity and 44% identity to the vitamin D3-25-hydroxylase (CYP27), whereas it has 42.6% similarity and 34% identity with the 25-hydroxyvitamin D3-24-hydroxylase (CYP24). Thus, it composes a new subfamily of the CYP27 family. Further, it is more closely related to the CYP27 than to the CYP24. The expression of P4501alpha mRNA was greatly increased in the kidney of vitamin D-deficient rats. In rats with the enhanced renal production of 1alpha,25-dihydroxyvitamin D3 (rats fed a low Ca diet), P4501alpha mRNA was greatly increased in the renal proximal convoluted tubules.
Figures



References
-
- DeLuca H F. FASEB J. 1988;2:224–236. - PubMed
-
- Suda T, Shinki T, Kurokawa K. Curr Opin Nephrol Hypertens. 1994;3:59–64. - PubMed
-
- Armbrecht H J, Okuda K, Wongsurawat N, Nemani R K, Chen M L, Boltz M A. J Steroid Biochem Mol Biol. 1992;43:1073–1081. - PubMed
-
- Okuda K, Usui E, Ohyama Y. J Lipid Res. 1995;36:1641–1652. - PubMed
-
- Nelson D R, Koymans L, Kamataki T, Stegeman J J, Feyereisen R, Waxman D J, Waterman M R, Gotoh O, Coon M J, Estabrook R W, Gunsalus I C, Nebert D W. Pharmacogenetics. 1996;6:1–42. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

