Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase
- PMID: 9371804
- PMCID: PMC24267
- DOI: 10.1073/pnas.94.24.13087
Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase
Abstract
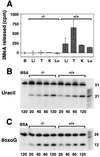
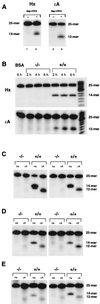
3-methyladenine (3MeA) DNA glycosylases remove 3MeAs from alkylated DNA to initiate the base excision repair pathway. Here we report the generation of mice deficient in the 3MeA DNA glycosylase encoded by the Aag (Mpg) gene. Alkyladenine DNA glycosylase turns out to be the major DNA glycosylase not only for the cytotoxic 3MeA DNA lesion, but also for the mutagenic 1,N6-ethenoadenine (epsilonA) and hypoxanthine lesions. Aag appears to be the only 3MeA and hypoxanthine DNA glycosylase in liver, testes, kidney, and lung, and the only epsilonA DNA glycosylase in liver, testes, and kidney; another epsilonA DNA glycosylase may be expressed in lung. Although alkyladenine DNA glycosylase has the capacity to remove 8-oxoguanine DNA lesions, it does not appear to be the major glycosylase for 8-oxoguanine repair. Fibroblasts derived from Aag -/- mice are alkylation sensitive, indicating that Aag -/- mice may be similarly sensitive.
Figures

Comment in
-
Life without DNA repair.Proc Natl Acad Sci U S A. 1997 Nov 25;94(24):12754-7. doi: 10.1073/pnas.94.24.12754. Proc Natl Acad Sci U S A. 1997. PMID: 9398071 Free PMC article. No abstract available.
References
-
- Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995.
-
- Lindahl T. Nature (London) 1976;259:64–66. - PubMed
-
- Chakravarti D, Ibeanu G C, Tano K, Mitra S. J Biol Chem. 1991;266:15710–15715. - PubMed
-
- O’Connor T R, Laval J. Biochem Biophys Res Commun. 1991;176:1170–1177. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

