Interleukin 9: a candidate gene for asthma
- PMID: 9371819
- PMCID: PMC24282
- DOI: 10.1073/pnas.94.24.13175
Interleukin 9: a candidate gene for asthma
Abstract
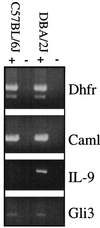
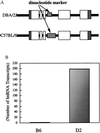
Asthma is a complex heritable inflammatory disorder of the airways associated with clinical signs of atopy and bronchial hyperresponsiveness. Recent studies localized a major gene for asthma to chromosome 5q31-q33 in humans. Thus, this segment of the genome represents a candidate region for genes that determine susceptibility to bronchial hyperresponsiveness and atopy in animal models. Homologs of candidate genes on human chromosome 5q31-q33 are found in four regions in the mouse genome, two on chromosome 18, and one each on chromosomes 11 and 13. We assessed bronchial responsiveness as a quantitative trait in mice and found it linked to chromosome 13. Interleukin 9 (IL-9) is located in the linked region and was analyzed as a gene candidate. The expression of IL-9 was markedly reduced in bronchial hyporesponsive mice, and the level of expression was determined by sequences within the qualitative trait locus (QTL). These data suggest a role for IL-9 in the complex pathogenesis of bronchial hyperresponsiveness as a risk factor for asthma.
Figures
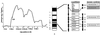

References
-
- McFadden E R, Jr, Gilbert I A. N Engl J Med. 1992;327:1928–1937. - PubMed
-
- Holgate S T, Church M K, Howarth P H, Morton N E, Frew A J, Ratko D. Int Arch Allergy Immunol. 1995;107:29–33. - PubMed
-
- Postma D S, Beecker E R, Amelung P J, Holroyd K J, Xu J, Panhuysen C I M, Meyers D A, Levitt R C. N Engl J Med. 1995;333:894–900. - PubMed
-
- The Collaborative Study on the Genetics of Asthma (CSGA) Nat Genet. 1997;15:389–392. - PubMed
-
- DeBry R W, Seldin M F. Genomics. 1996;33:337–351. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

