The ARS309 chromosomal replicator of Saccharomyces cerevisiae depends on an exceptional ARS consensus sequence
- PMID: 9380711
- PMCID: PMC23486
- DOI: 10.1073/pnas.94.20.10786
The ARS309 chromosomal replicator of Saccharomyces cerevisiae depends on an exceptional ARS consensus sequence
Abstract
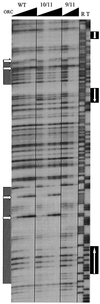
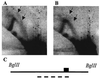
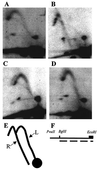
Autonomously replicating sequence (ARS) elements, which function as the cis-acting chromosomal replicators in the yeast Saccharomyces cerevisiae, depend upon an essential copy of the 11-bp ARS consensus sequence (ACS) for activity. Analysis of the chromosome III replicator ARS309 unexpectedly revealed that its essential ACS differs from the canonical ACS at two positions. One of the changes observed in ARS309 inactivates other ARS elements. This atypical ACS binds the origin recognition complex efficiently and is required for chromosomal replication origin activity. Comparison of the essential ACS of ARS309 with the essential regions of other ARS elements revealed an expanded 17-bp conserved sequence that efficiently predicts the essential core of ARS elements.
Figures


Similar articles
-
Analysis of chromosome III replicators reveals an unusual structure for the ARS318 silencer origin and a conserved WTW sequence within the origin recognition complex binding site.Mol Cell Biol. 2008 Aug;28(16):5071-81. doi: 10.1128/MCB.00206-08. Epub 2008 Jun 23. Mol Cell Biol. 2008. PMID: 18573888 Free PMC article.
-
Functional conservation of multiple elements in yeast chromosomal replicators.Mol Cell Biol. 1994 Nov;14(11):7643-51. doi: 10.1128/mcb.14.11.7643-7651.1994. Mol Cell Biol. 1994. PMID: 7935478 Free PMC article.
-
Multiple DNA elements in ARS305 determine replication origin activity in a yeast chromosome.Nucleic Acids Res. 1996 Mar 1;24(5):816-23. doi: 10.1093/nar/24.5.816. Nucleic Acids Res. 1996. PMID: 8600446 Free PMC article.
-
Structure, replication efficiency and fragility of yeast ARS elements.Res Microbiol. 2012 May;163(4):243-53. doi: 10.1016/j.resmic.2012.03.003. Epub 2012 Mar 28. Res Microbiol. 2012. PMID: 22504206 Review.
-
The dual role of autonomously replicating sequences as origins of replication and as silencers.Curr Genet. 2009 Aug;55(4):357-63. doi: 10.1007/s00294-009-0265-7. Epub 2009 Jul 26. Curr Genet. 2009. PMID: 19633981 Review.
Cited by
-
Roles for internal and flanking sequences in regulating the activity of mating-type-silencer-associated replication origins in Saccharomyces cerevisiae.Genetics. 2001 Sep;159(1):35-45. doi: 10.1093/genetics/159.1.35. Genetics. 2001. PMID: 11560885 Free PMC article.
-
Mcm1 promotes replication initiation by binding specific elements at replication origins.Mol Cell Biol. 2004 Jul;24(14):6514-24. doi: 10.1128/MCB.24.14.6514-6524.2004. Mol Cell Biol. 2004. PMID: 15226450 Free PMC article.
-
The requirement of yeast replication origins for pre-replication complex proteins is modulated by transcription.Nucleic Acids Res. 2005 Apr 28;33(8):2410-20. doi: 10.1093/nar/gki539. Print 2005. Nucleic Acids Res. 2005. PMID: 15860777 Free PMC article.
-
Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae.BMC Genomics. 2006 Oct 26;7:276. doi: 10.1186/1471-2164-7-276. BMC Genomics. 2006. PMID: 17067396 Free PMC article.
-
Ordered and disordered regions of the Origin Recognition Complex direct differential in vivo binding at distinct motif sequences.Nucleic Acids Res. 2024 Jun 10;52(10):5720-5731. doi: 10.1093/nar/gkae249. Nucleic Acids Res. 2024. PMID: 38597680 Free PMC article.
References
-
- Theis J F, Newlon C S. In: The Mycota. Brambl R, Marzluf G A, editors. Vol. 3. Berlin: Springer; 1996. pp. 1–28.
-
- Stinchcomb D T, Struhl K, Davis R W. Nature (London) 1979;282:39–43. - PubMed
-
- Huberman J A, Spotila L D, Nawotka K A, el-Assouli S M, Davis L R. Cell. 1987;51:473–481. - PubMed
-
- Brewer B J, Fangman W L. Cell. 1987;51:463–471. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

