Stu2p: A microtubule-binding protein that is an essential component of the yeast spindle pole body
- PMID: 9382872
- PMCID: PMC2140218
- DOI: 10.1083/jcb.139.5.1271
Stu2p: A microtubule-binding protein that is an essential component of the yeast spindle pole body
Abstract
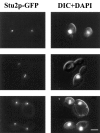
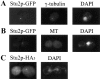
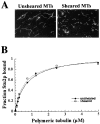
Previously we isolated tub2-423, a cold-sensitive allele of the Saccharomyces cerevisiae gene encoding beta-tubulin that confers a defect in mitotic spindle function. In an attempt to identify additional proteins that are important for spindle function, we screened for suppressors of the cold sensitivity of tub2-423 and obtained two alleles of a novel gene, STU2. STU2 is an essential gene and encodes a protein whose sequence is similar to proteins identified in a variety of organisms. Stu2p localizes primarily to the spindle pole body (SPB) and to a lesser extent along spindle microtubules. Localization to the SPB is not dependent on the presence of microtubules, indicating that Stu2p is an integral component of the SPB. Stu2p also binds microtubules in vitro. We have localized the microtubule-binding domain of Stu2p to a highly basic 100-amino acid region. This region contains two imperfect repeats; both repeats appear to contribute to microtubule binding to similar extents. These results suggest that Stu2p may play a role in the attachment, organization, and/or dynamics of microtubule ends at the SPB.
Figures




References
-
- Biernat J, Gustke N, Drewes G, Mandelkow EM, Mandelkow E. Phosphorylation of Ser262 strongly reduces binding of tau to microtubules: distinction between PHF-like immunoreactivity and microtubule binding. Neuron. 1993;11:153–163. - PubMed
-
- Bramblett GT, Goedert M, Jakes R, Merrick SE, Trojanowski JQ, Lee VM. Abnormal tau phosphorylation at Ser396 in Alzheimer's disease recapitulates development and contributes to reduced microtubule binding. Neuron. 1993;10:1089–1099. - PubMed
-
- Byers, B. 1981. Cytology of the yeast life cycle. In The Molecular Biology of the Yeast Saccharomyces. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York. 59–96.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

