Aldehyde dehydrogenase class 3 expression: identification of a cornea-preferred gene promoter in transgenic mice
- PMID: 9391071
- PMCID: PMC28351
- DOI: 10.1073/pnas.94.25.13594
Aldehyde dehydrogenase class 3 expression: identification of a cornea-preferred gene promoter in transgenic mice
Abstract
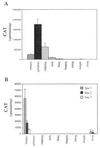
Aldehyde dehydrogenase class 3 (ALDH3) constitutes 20-40% of the total water-soluble proteins in the mammalian cornea. Here, we show by Northern blot analysis that ALDH3 expression in the mouse is at least 500-fold higher in the cornea than in any other tissue examined, with very low levels of expression detected in the stomach, urinary bladder, ocular lens, and lung. Histochemical localization reveals that this exceptional level of expression in the mouse cornea occurs in the anterior epithelial cells and that little ALDH3 is present in the keratocytes or corneal endothelial cells. A 13-kbp mouse ALDH3 promoter fragment containing >12 kbp of the 5' flanking sequence, the 40-bp untranslated first exon, and 29 bp of intron 1 directed cat reporter gene expression to tissues that express the endogenous ALDH3 gene, except that transgene promoter activity was higher in the stomach and bladder than in the cornea. By contrast, when driven by a 4.4-kbp mouse ALDH3 promoter fragment [1,050-bp 5' flanking region, exon 1, intron 1 (3.4 kbp), and 7 bp of exon 2] expression of the cat reporter gene was confined to the corneal epithelial cells, except for very low levels in the liver, effectively reproducing the corneal expression pattern of the endogenous ALDH3 gene. These results indicate that tissue-specific expression of ALDH3 is determined by positive and negative elements in the 5' flanking region of the gene and suggests putative silencers located in intron 1. We demonstrate regulatory sequences capable of directing cornea-specific gene expression, affording the opportunity for genetic engineering in this transparent tissue.
Figures



References
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

