Molecular evolution of two vertebrate aryl hydrocarbon (dioxin) receptors (AHR1 and AHR2) and the PAS family
- PMID: 9391097
- PMCID: PMC28377
- DOI: 10.1073/pnas.94.25.13743
Molecular evolution of two vertebrate aryl hydrocarbon (dioxin) receptors (AHR1 and AHR2) and the PAS family
Abstract
The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor through which halogenated aromatic hydrocarbons such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) cause altered gene expression and toxicity. The AHR belongs to the basic helix-loop-helix/Per-ARNT-Sim (bHLH-PAS) family of transcriptional regulatory proteins, whose members play key roles in development, circadian rhythmicity, and environmental homeostasis; however, the normal cellular function of the AHR is not yet known. As part of a phylogenetic approach to understanding the function and evolutionary origin of the AHR, we sequenced the PAS homology domain of AHRs from several species of early vertebrates and performed phylogenetic analyses of these AHR amino acid sequences in relation to mammalian AHRs and 24 other members of the PAS family. AHR sequences were identified in a teleost (the killifish Fundulus heteroclitus), two elasmobranch species (the skate Raja erinacea and the dogfish Mustelus canis), and a jawless fish (the lamprey Petromyzon marinus). Two putative AHR genes, designated AHR1 and AHR2, were found both in Fundulus and Mustelus. Phylogenetic analyses indicate that the AHR2 genes in these two species are orthologous, suggesting that an AHR gene duplication occurred early in vertebrate evolution and that multiple AHR genes may be present in other vertebrates. Database searches and phylogenetic analyses identified four putative PAS proteins in the nematode Caenorhabditis elegans, including possible AHR and ARNT homologs. Phylogenetic analysis of the PAS gene family reveals distinct clades containing both invertebrate and vertebrate PAS family members; the latter include paralogous sequences that we propose have arisen by gene duplication early in vertebrate evolution. Overall, our analyses indicate that the AHR is a phylogenetically ancient protein present in all living vertebrate groups (with a possible invertebrate homolog), thus providing an evolutionary perspective to the study of dioxin toxicity and AHR function.
Figures

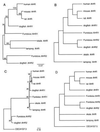

References
-
- Poland A, Knutson J C. Annu Rev Pharmacol Toxicol. 1982;22:517–554. - PubMed
-
- Hoffman E C, Reyes H, Chu F-F, Sander F, Conley L H, Brooks B A, Hankinson O. Science. 1991;252:954–958. - PubMed
-
- Hankinson O. Annu Rev Pharmacol Toxicol. 1995;35:307–340. - PubMed
-
- Schmidt J V, Bradfield C A. Annu Rev Cell Dev Biol. 1996;12:55–89. - PubMed
-
- Nambu J R, Lewis J O, Wharton K A, Crews S T. Cell. 1991;67:1157–1167. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

