Did homeodomain proteins duplicate before the origin of angiosperms, fungi, and metazoa?
- PMID: 9391098
- PMCID: PMC28378
- DOI: 10.1073/pnas.94.25.13749
Did homeodomain proteins duplicate before the origin of angiosperms, fungi, and metazoa?
Abstract
Homeodomain proteins are transcription factors that play a critical role in early development in eukaryotes. These proteins previously have been classified into numerous subgroups whose phylogenetic relationships are unclear. Our phylogenetic analysis of representative eukaryotic sequences suggests that there are two major groups of homeodomain proteins, each containing sequences from angiosperms, metazoa, and fungi. This result, based on parsimony and neighbor-joining analyses of primary amino acid sequences, was supported by two additional features of the proteins. The two protein groups are distinguished by an insertion/deletion in the homeodomain, between helices I and II. In addition, an amphipathic alpha-helical secondary structure in the region N terminal of the homeodomain is shared by angiosperm and metazoan sequences in one group. These results support the hypothesis that there was at least one duplication of homeobox genes before the origin of angiosperms, fungi, and metazoa. This duplication, in turn, suggests that these proteins had diverse functions early in the evolution of eukaryotes. The shared secondary structure in angiosperm and metazoan sequences points to an ancient conserved functional domain.
Figures
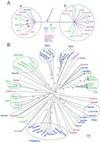


References
-
- McGinnis W, Levine M S, Hafen E, Kuroiwa A, Gehring W J. Nature (London) 1984;308:428–433. - PubMed
-
- Scott M P, Tamkun J W, Hartzell G W. Biochim Biophys Acta. 1989;989:25–48. - PubMed
-
- Kappen C, Schughart K, Ruddle F H. Genomics. 1993;18:54–70. - PubMed
-
- Bürglin T R. In: Guidebook to the Homeobox Genes. Duboule D, editor. New York: Oxford; 1994. pp. 27–71.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

