Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe
- PMID: 9391101
- PMCID: PMC28381
- DOI: 10.1073/pnas.94.25.13765
Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe
Abstract
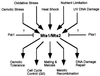
Homologous recombination hotspots increase the frequency of recombination in nearby DNA. The M26 hotspot in the ade6 gene of Schizosaccharomyces pombe is a meiotic hotspot with a discrete, cis-acting nucleotide sequence (5'-ATGACGT-3') defined by extensive mutagenesis. A heterodimeric M26 DNA binding protein, composed of subunits Mts1 and Mts2, has been identified and purified 40,000-fold. Cloning, disruption, and genetic analyses of the mts genes demonstrate that the Mts1/Mts2 heterodimer is essential for hotspot activity. This provides direct evidence that a specific trans-acting factor, binding to a cis-acting site with a unique nucleotide sequence, is required to activate this meiotic hotspot. Intriguingly, the Mts1/Mts2 protein subunits are identical to the recently described transcription factors Atf1 (Gad7) and Pcr1, which are required for a variety of stress responses. However, we report differential dependence on the Mts proteins for hotspot activation and stress response, suggesting that these proteins are multifunctional and have distinct activities. Furthermore, ade6 mRNA levels are equivalent in hotspot and nonhotspot meioses and do not change in mts mutants, indicating that hotspot activation is not a consequence of elevated transcription levels. These findings suggest an intimate but separable link between the regulation of transcription and meiotic recombination. Other studies have recently shown that the Mts1/Mts2 protein and M26 sites are involved in meiotic recombination elsewhere in the S. pombe genome, suggesting that these factors help regulate the timing and distribution of homologous recombination.
Figures




Comment in
-
Relationship between transcription and initiation of meiotic recombination: toward chromatin accessibility.Proc Natl Acad Sci U S A. 1998 Jan 6;95(1):87-9. doi: 10.1073/pnas.95.1.87. Proc Natl Acad Sci U S A. 1998. PMID: 9419331 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

