Mitochondrial mutational spectra in human cells and tissues
- PMID: 9391107
- PMCID: PMC28387
- DOI: 10.1073/pnas.94.25.13798
Mitochondrial mutational spectra in human cells and tissues
Abstract
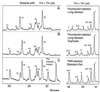
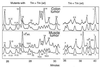
We have found that human organs such as colon, lung, and muscle, as well as their derived tumors, share nearly all mitochondrial hotspot point mutations. Seventeen hotspots, primarily G --> A and A --> G transitions, have been identified in the mitochondrial sequence of base pairs 10,030-10,130. Mutant fractions increase with the number of cell generations in a human B cell line, TK6, indicating that they are heritable changes. The mitochondrial point mutation rate appears to be more than two orders of magnitude higher than the nuclear point mutation rate in TK6 cells and in human tissues. The similarity of the hotspot sets in vivo and in vitro leads us to conclude that human mitochondrial point mutations in the sequence studied are primarily spontaneous in origin and arise either from DNA replication error or reactions of DNA with endogenous metabolites. The predominance of transition mutations and the high number of hotspots in this short sequence resembles spectra produced by DNA polymerases in vitro.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

