Us9, a stable lysine-less herpes simplex virus 1 protein, is ubiquitinated before packaging into virions and associates with proteasomes
- PMID: 9391137
- PMCID: PMC28417
- DOI: 10.1073/pnas.94.25.13973
Us9, a stable lysine-less herpes simplex virus 1 protein, is ubiquitinated before packaging into virions and associates with proteasomes
Abstract
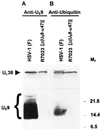
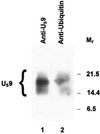
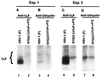
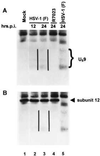
The US9 gene of herpes simplex virus 1 encodes a virion tegument protein with a predicted Mr of 10,000. Earlier studies have shown that the gene is not essential for viral replication in cells in culture. We report that (i) US9 forms in denaturing polyacrylamide gels multiple overlapping bands ranging in Mr from 12,000 to 25,000; (ii) the protein recovered from infected cells or purified virions reacts with anti-ubiquitin antibodies; (iii) autoradiographic images of US9 protein immunoprecipitated from cells infected with [35S]methionine-labeled virus indicate that the protein is stable for at least 4 h after entry into cells (the protein was also stable for at least 4 h after a 1-h labeling interval 12 h after infection); (iv) antibody to subunit 12 of proteasomes pulls down US9 protein from herpes simplex virus-infected cell lysates; and (v) the US9 gene is highly conserved among the members of the alpha subfamily of herpes viruses, and the US9 gene product lacks lysines. We conclude that US9 is a lysine-less, ubiquitinated protein that interacts with the ubiquitin-dependent pathway for degradation of proteins and that this function may be initiated at the time of entry of the virus into the cell.
Figures


References
-
- McGeoch D J, Dolan A, Donald S, Rixon F J. J Mol Biol. 1985;181:1–13. - PubMed
-
- Frame M C, McGeoch D J, Rixon F J, Orr A C, Marsden H S. Virology. 1986;150:321–332. - PubMed
-
- Roizman B, Sears A E. In: Fields’ Virology. 3rd Ed. Fields B N, Knipe D M, Howley P, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. New York: Raven; 1996. pp. 2231–2295.
-
- Jentsch F. Annu Rev Genet. 1992;26:179–207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

