Dissociation of Oct-1 from the nuclear peripheral structure induces the cellular aging-associated collagenase gene expression
- PMID: 9398664
- PMCID: PMC25716
- DOI: 10.1091/mbc.8.12.2407
Dissociation of Oct-1 from the nuclear peripheral structure induces the cellular aging-associated collagenase gene expression
Abstract
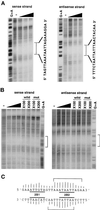
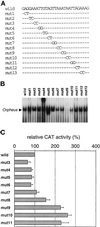
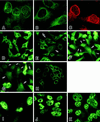
The cellular aging-associated transcriptional repressor that we previously named as Orpheus was identical to Oct-1, a member of the POU domain family. Oct-1 represses the collagenase gene, one of the cellular aging-associated genes, by interacting with an AT-rich cis-element in the upstream of the gene in preimmortalized cells at earlier population-doubling levels and in immortalized cells. In these stages of cells, considerable fractions of the Oct-1 protein were prominently localized in the nuclear periphery and colocalized with lamin B. During the cellular aging process, however, this subspecies of Oct-1 disappeared from the nuclear periphery. The cells lacking the nuclear peripheral Oct-1 protein exhibited strong collagenase expression and carried typical senescent morphologies. Concomitantly, the binding activity and the amount of nuclear Oct-1 protein were reduced in the aging process and resumed after immortalization. However, the whole cellular amounts of Oct-1 protein were not significantly changed during either process. Thus, the cellular aging-associated genes including the collagenase gene seemed to be derepressed by the dissociation of Oct-1 protein from the nuclear peripheral structure. Oct-1 may form a transcriptional repressive apparatus by anchoring nuclear matrix attachment regions onto the nuclear lamina in the nuclear periphery.
Figures



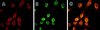


Similar articles
-
Silencing mediator for retinoid and thyroid hormone receptors interacts with octamer transcription factor-1 and acts as a transcriptional repressor.J Biol Chem. 2001 Mar 30;276(13):9720-5. doi: 10.1074/jbc.M008531200. Epub 2000 Dec 27. J Biol Chem. 2001. PMID: 11134019
-
Immortalization-susceptible elements and their binding factors mediate rejuvenation of regulation of the type I collagenase gene in simian virus 40 large T antigen-transformed immortal human fibroblasts.Mol Cell Biol. 1994 Nov;14(11):7182-94. doi: 10.1128/mcb.14.11.7182-7194.1994. Mol Cell Biol. 1994. PMID: 7935433 Free PMC article.
-
DNA-dependent conversion of Oct-1 and Oct-2 into transcriptional repressors by Groucho/TLE.Nucleic Acids Res. 2005 Aug 15;33(14):4618-25. doi: 10.1093/nar/gki744. Print 2005. Nucleic Acids Res. 2005. PMID: 16103132 Free PMC article.
-
The virtuoso of versatility: POU proteins that flex to fit.J Mol Biol. 2000 Oct 6;302(5):1023-39. doi: 10.1006/jmbi.2000.4107. J Mol Biol. 2000. PMID: 11183772 Review.
-
Regulation of transcription factor activity during cellular aging.Biochem Cell Biol. 1996;74(4):523-34. doi: 10.1139/o96-056. Biochem Cell Biol. 1996. PMID: 8960358 Review.
Cited by
-
Regulation of Oct1/Pou2f1 transcription activity by O-GlcNAcylation.FASEB J. 2013 Jul;27(7):2807-17. doi: 10.1096/fj.12-220897. Epub 2013 Apr 11. FASEB J. 2013. PMID: 23580612 Free PMC article.
-
The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation.Genes Dev. 2006 Jun 1;20(11):1447-57. doi: 10.1101/gad.1419506. Epub 2006 May 16. Genes Dev. 2006. PMID: 16705039 Free PMC article.
-
Accumulation of multipotent progenitors with a basal differentiation bias during aging of human mammary epithelia.Cancer Res. 2012 Jul 15;72(14):3687-701. doi: 10.1158/0008-5472.CAN-12-0157. Epub 2012 May 2. Cancer Res. 2012. PMID: 22552289 Free PMC article.
-
The inner nuclear envelope as a transcription factor resting place.EMBO Rep. 2007 Oct;8(10):914-9. doi: 10.1038/sj.embor.7401075. EMBO Rep. 2007. PMID: 17906672 Free PMC article. Review.
-
Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression.J Cell Biol. 2007 Feb 26;176(5):593-603. doi: 10.1083/jcb.200607054. Epub 2007 Feb 20. J Cell Biol. 2007. PMID: 17312019 Free PMC article.
References
-
- Bauer EA, Silverman N, Busiek DF, Kronberger A, Deuel TF. Diminished response of Werner’s syndrome fibroblasts to growth factors PDGF and FGF. Science. 1986;234:1240–1243. - PubMed
-
- Bendall AJ, Sturm RA, Danoy PAC, Molloy PL. Broad binding-site specificity and affinity properties of octamer 1 and brain octamer-binding proteins. Eur J Biochem. 1993;217:799–811. - PubMed
-
- Burke EM, Horton WE, Pearson JD, Crow MT, Martin GR. Altered transcriptional regulation of human interstitial collagenase in cultured skin fibroblasts from older donors. Exp Gerontol. 1994;29:37–53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

