Characterization of Saccharomyces cerevisiae dna2 mutants suggests a role for the helicase late in S phase
- PMID: 9398673
- PMCID: PMC25725
- DOI: 10.1091/mbc.8.12.2519
Characterization of Saccharomyces cerevisiae dna2 mutants suggests a role for the helicase late in S phase
Abstract
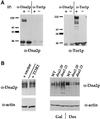
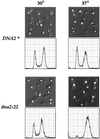
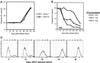
The TOR proteins, originally identified as targets of the immunosuppressant rapamycin, contain an ATM-like "lipid kinase" domain and are required for early G1 progression in eukaryotes. Using a screen to identify Saccharomyces cerevisiae mutants requiring overexpression of Tor1p for viability, we have isolated mutations in a gene we call ROT1 (requires overexpression of Tor1p). This gene is identical to DNA2, encoding a helicase required for DNA replication. As with its role in cell cycle progression, both the N-terminal and C-terminal regions, as well as the kinase domain of Tor1p, are required for rescue of dna2 mutants. Dna2 mutants are also rescued by Tor2p and show synthetic lethality with tor1 deletion mutants under specific conditions. Temperature-sensitive (Ts) dna2 mutants arrest irreversibly at G2/M in a RAD9- and MEC1-dependent manner, suggesting that Dna2p has a role in S phase. Frequencies of mitotic recombination and chromosome loss are elevated in dna2 mutants, also supporting a role for the protein in DNA synthesis. Temperature-shift experiments indicate that Dna2p functions during late S phase, although dna2 mutants are not deficient in bulk DNA synthesis. These data suggest that Dna2p is not required for replication fork progression but may be needed for a later event such as Okazaki fragment maturation.
Figures
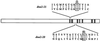






References
-
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Biswas EE, Chen PH, Leszyk J, Biswas SB. Biochemical and genetic characterization of a replication protein A dependent DNA helicase from the yeast, Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;206:850–856. - PubMed
-
- Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

