An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae
- PMID: 9405607
- PMCID: PMC24951
- DOI: 10.1073/pnas.94.26.14300
An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae
Abstract
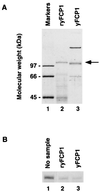
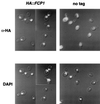
One of the essential components of a phosphatase that specifically dephosphorylates the Saccharomyces cerevisiae RNA polymerase II (RPII) large subunit C-terminal domain (CTD) is a novel polypeptide encoded by an essential gene termed FCP1. The Fcp1 protein is localized to the nucleus, and it binds the largest subunit of the yeast general transcription factor IIF (Tfg1). In vitro, transcription factor IIF stimulates phosphatase activity in the presence of Fcp1 and a second complementing fraction. Two distinct regions of Fcp1 are capable of binding to Tfg1, but the C-terminal Tfg1 binding domain is dispensable for activity in vivo and in vitro. Sequence comparison reveals that residues 173-357 of Fcp1 correspond to an amino acid motif present in proteins of unknown function predicted in many organisms.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

