Thermodynamic stability of wild-type and mutant p53 core domain
- PMID: 9405613
- PMCID: PMC24967
- DOI: 10.1073/pnas.94.26.14338
Thermodynamic stability of wild-type and mutant p53 core domain
Abstract
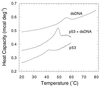
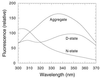
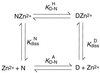
Some 50% of human cancers are associated with mutations in the core domain of the tumor suppressor p53. Many mutations are thought just to destabilize the protein. To assess this and the possibility of rescue, we have set up a system to analyze the stability of the core domain and its mutants. The use of differential scanning calorimetry or spectroscopy to measure its melting temperature leads to irreversible denaturation and aggregation and so is useful as only a qualitative guide to stability. There are excellent two-state denaturation curves on the addition of urea that may be analyzed quantitatively. One Zn2+ ion remains tightly bound in the holo-form of p53 throughout the denaturation curve. The stability of wild type is 6.0 kcal (1 kcal = 4.18 kJ)/mol at 25 degrees C and 9.8 kcal/mol at 10 degrees C. The oncogenic mutants R175H, C242S, R248Q, R249S, and R273H are destabilized by 3.0, 2.9, 1.9, 1.9, and 0.4 kcal/mol, respectively. Under certain denaturing conditions, the wild-type domain forms an aggregate that is relatively highly fluorescent at 340 nm on excitation at 280 nm. The destabilized mutants give this fluorescence under milder denaturation conditions.
Figures


References
-
- Lane D P. Nature (London) 1992;358:15–16. - PubMed
-
- Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Cancer Res. 1991;51:6304–6311. - PubMed
-
- Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. Nature (London) 1997;389:300–305. - PubMed
-
- Ciechanover A, Shkedy D, Oren M, Bercovich B. J Biol Chem. 1994;269:9582–9589. - PubMed
-
- Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

