Archaeal-type lysyl-tRNA synthetase in the Lyme disease spirochete Borrelia burgdorferi
- PMID: 9405621
- PMCID: PMC24988
- DOI: 10.1073/pnas.94.26.14383
Archaeal-type lysyl-tRNA synthetase in the Lyme disease spirochete Borrelia burgdorferi
Abstract
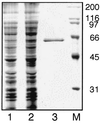
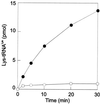
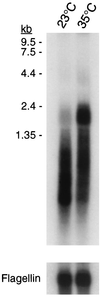
Lysyl-tRNAs are essential for protein biosynthesis by ribosomal mRNA translation in all organisms. They are synthesized by lysyl-tRNA synthetases (EC 6.1.1.6), a group of enzymes composed of two unrelated families. In bacteria and eukarya, all known lysyl-tRNA synthetases are subclass IIc-type aminoacyl-tRNA synthetases, whereas some archaea have been shown to contain an unrelated class I-type lysyl-tRNA synthetase. Examination of the preliminary genomic sequence of the bacterial pathogen Borrelia burgdorferi, the causative agent of Lyme disease, indicated the presence of an open reading frame with over 55% similarity at the amino acid level to archaeal class I-type lysyl-tRNA synthetases. In contrast, no coding region with significant similarity to any class II-type lysyl-tRNA synthetase could be detected. Heterologous expression of this open reading frame in Escherichia coli led to the production of a protein with canonical lysyl-tRNA synthetase activity in vitro. Analysis of B. burgdorferi mRNA showed that the lysyl-tRNA synthetase-encoding gene is highly expressed, confirming that B. burgdorferi contains a functional class I-type lysyl-tRNA synthetase. The detection of an archaeal-type lysyl-tRNA synthetase in B. burgdorferi and other pathogenic spirochetes, but not to date elsewhere in bacteria or eukarya, indicates that the gene that encodes this enzyme has a common origin with its orthologue from the archaeal kingdom. This difference between the lysyl-tRNA synthetases of spirochetes and their hosts may be readily exploitable for the development of anti-spirochete therapeutics.
Figures




Similar articles
-
Context-dependent anticodon recognition by class I lysyl-tRNA synthetases.Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14224-8. doi: 10.1073/pnas.97.26.14224. Proc Natl Acad Sci U S A. 2000. PMID: 11121028 Free PMC article.
-
Transfer RNA recognition by class I lysyl-tRNA synthetase from the Lyme disease pathogen Borrelia burgdorferi.FEBS Lett. 2005 May 9;579(12):2629-34. doi: 10.1016/j.febslet.2005.04.001. Epub 2005 Apr 9. FEBS Lett. 2005. PMID: 15862301
-
Differentiation of Borrelia burgdorferi sensu lato strains using class I lysyl-tRNA synthetase-encoding genes.Med Microbiol Immunol. 2003 May;192(2):79-83. doi: 10.1007/s00430-002-0149-7. Epub 2002 Sep 25. Med Microbiol Immunol. 2003. PMID: 12736820
-
Lysyl-tRNA synthetase.Biol Chem Hoppe Seyler. 1995 Aug;376(8):451-72. doi: 10.1515/bchm3.1995.376.8.451. Biol Chem Hoppe Seyler. 1995. PMID: 7576245 Review.
-
Archaeal aminoacyl-tRNA synthesis: diversity replaces dogma.Genetics. 1999 Aug;152(4):1269-76. doi: 10.1093/genetics/152.4.1269. Genetics. 1999. PMID: 10430557 Free PMC article. Review.
Cited by
-
Context-dependent anticodon recognition by class I lysyl-tRNA synthetases.Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14224-8. doi: 10.1073/pnas.97.26.14224. Proc Natl Acad Sci U S A. 2000. PMID: 11121028 Free PMC article.
-
In silico detection of tRNA sequence features characteristic to aminoacyl-tRNA synthetase class membership.Nucleic Acids Res. 2007;35(16):5593-609. doi: 10.1093/nar/gkm598. Epub 2007 Aug 17. Nucleic Acids Res. 2007. PMID: 17704131 Free PMC article.
-
Structures of two bacterial resistance factors mediating tRNA-dependent aminoacylation of phosphatidylglycerol with lysine or alanine.Proc Natl Acad Sci U S A. 2015 Aug 25;112(34):10691-6. doi: 10.1073/pnas.1511167112. Epub 2015 Aug 10. Proc Natl Acad Sci U S A. 2015. PMID: 26261323 Free PMC article.
-
The T box regulatory element controlling expression of the class I lysyl-tRNA synthetase of Bacillus cereus strain 14579 is functional and can be partially induced by reduced charging of asparaginyl-tRNAAsn.BMC Microbiol. 2010 Jul 22;10:196. doi: 10.1186/1471-2180-10-196. BMC Microbiol. 2010. PMID: 20649968 Free PMC article.
-
Non-homologous isofunctional enzymes: a systematic analysis of alternative solutions in enzyme evolution.Biol Direct. 2010 Apr 30;5:31. doi: 10.1186/1745-6150-5-31. Biol Direct. 2010. PMID: 20433725 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

