Plasmon analyses of Triticum (wheat) and Aegilops: PCR-single-strand conformational polymorphism (PCR-SSCP) analyses of organellar DNAs
- PMID: 9405654
- PMCID: PMC25058
- DOI: 10.1073/pnas.94.26.14570
Plasmon analyses of Triticum (wheat) and Aegilops: PCR-single-strand conformational polymorphism (PCR-SSCP) analyses of organellar DNAs
Abstract
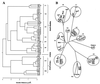
To investigate phylogenetic relationships among plasmons in Triticum and Aegilops, PCR-single-strand conformational polymorphism (PCR-SSCP) analyses were made of 14.0-kb chloroplast (ct) and 13. 7-kb mitochondrial (mt)DNA regions that were isolated from 46 alloplasmic wheat lines and one euplasmic line. These plasmons represent 31 species of the two genera. The ct and mtDNA regions included 10 and 9 structural genes, respectively. A total of 177 bands were detected, of which 40.6% were variable. The proportion of variable bands in ctDNA (51.1%) was higher than that of mtDNA (28. 9%). The phylogenetic trees of plasmons, derived by two different models, indicate a common picture of plasmon divergence in the two genera and suggest three major groups of plasmons (Einkorn, Triticum, and Aegilops). Because of uniparental plasmon transmission, the maternal parents of all but one polyploid species were identified. Only one Aegilops species, Ae. speltoides, was included in the Triticum group, suggesting that this species is the plasmon and B and G genome donor of all polyploid wheats. ctDNA variations were more intimately correlated with vegetative characters, whereas mtDNA variations were more closely correlated with reproductive characters. Plasmon divergence among the diploids of the two genera largely paralleled genome divergence. The relative times of origin of the polyploid species were inferred from genetic distances from their putative maternal parents.
Figures


References
-
- Kihara H. Cytologia. 1951;16:177–193.
-
- Tsunewaki K. In: Methods of Genome Analysis in Plants. Jauhar P P, editor. Boca Raton, FL: CRC; 1996. pp. 271–298.
-
- Vedel F, Quetier F, Dosba F, Doussinault G. Plant Sci Lett. 1978;13:97–102.
-
- Bowman C M, Koller B, Delius H, Dyer T A. Mol Gen Genet. 1981;183:93–101.
-
- Ogihara Y, Tsunewaki K. Theor Appl Genet. 1988;76:321–332. - PubMed
LinkOut - more resources
Full Text Sources

