Comparative mapping of the human 22q11 chromosomal region and the orthologous region in mice reveals complex changes in gene organization
- PMID: 9405660
- PMCID: PMC25069
- DOI: 10.1073/pnas.94.26.14608
Comparative mapping of the human 22q11 chromosomal region and the orthologous region in mice reveals complex changes in gene organization
Abstract
The region of human chromosome 22q11 is prone to rearrangements. The resulting chromosomal abnormalities are involved in Velo-cardio-facial and DiGeorge syndromes (VCFS and DGS) (deletions), "cat eye" syndrome (duplications), and certain types of tumors (translocations). As a prelude to the development of mouse models for VCFS/DGS by generating targeted deletions in the mouse genome, we examined the organization of genes from human chromosome 22q11 in the mouse. Using genetic linkage analysis and detailed physical mapping, we show that genes from a relatively small region of human 22q11 are distributed on three mouse chromosomes (MMU6, MMU10, and MMU16). Furthermore, although the region corresponding to about 2.5 megabases of the VCFS/DGS critical region is located on mouse chromosome 16, the relative organization of the region is quite different from that in humans. Our results show that the instability of the 22q11 region is not restricted to humans but may have been present throughout evolution. The results also underscore the importance of detailed comparative mapping of genes in mice and humans as a prerequisite for the development of mouse models of human diseases involving chromosomal rearrangements.
Figures
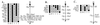


References
-
- Biegel J A, Allen C S, Kawasaki K, Shimizu N, Budarf M L, Bell C J. Gene Chromosomes Cancer. 1996;16:94–105. - PubMed
-
- Iselius L, Lindsten J, Aurias A, Fraccaro M, Bastard C, Bottelli A M, Bui T H, Caufin D, Dalpra L, Delendi N, et al. Hum Genet. 1983;64:343–355. - PubMed
-
- Scambler P J, Kelly D, Lindsay E, Williamson R, Goldberg R, Shprintzen R, Wilson D I, Goodship J A, Cross I E, Burn J. Lancet. 1992;339:1138–1139. - PubMed
-
- Kelly D, Goldberg R, Wilson D, Lindsay E, Carey A, Goodship J, Burn J, Cross I, Shprintzen R J, Scambler P J. Am J Med Genet. 1993;45:308–312. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

