The anticoagulant activation of antithrombin by heparin
- PMID: 9405673
- PMCID: PMC25092
- DOI: 10.1073/pnas.94.26.14683
The anticoagulant activation of antithrombin by heparin
Abstract
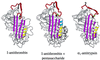
Antithrombin, a plasma serpin, is relatively inactive as an inhibitor of the coagulation proteases until it binds to the heparan side chains that line the microvasculature. The binding specifically occurs to a core pentasaccharide present both in the heparans and in their therapeutic derivative heparin. The accompanying conformational change of antithrombin is revealed in a 2.9-A structure of a dimer of latent and active antithrombins, each in complex with the high-affinity pentasaccharide. Inhibitory activation results from a shift in the main sheet of the molecule from a partially six-stranded to a five-stranded form, with extrusion of the reactive center loop to give a more exposed orientation. There is a tilting and elongation of helix D with the formation of a 2-turn helix P between the C and D helices. Concomitant conformational changes at the heparin binding site explain both the initial tight binding of antithrombin to the heparans and the subsequent release of the antithrombin-protease complex into the circulation. The pentasaccharide binds by hydrogen bonding of its sulfates and carboxylates to Arg-129 and Lys-125 in the D-helix, to Arg-46 and Arg-47 in the A-helix, to Lys-114 and Glu-113 in the P-helix, and to Lys-11 and Arg-13 in a cleft formed by the amino terminus. This clear definition of the binding site will provide a structural basis for developing heparin analogues that are more specific toward their intended target antithrombin and therefore less likely to exhibit side effects.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

