Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome
- PMID: 9405686
- PMCID: PMC25110
- DOI: 10.1073/pnas.94.26.14759
Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome
Abstract
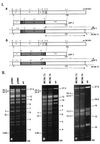
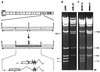
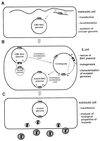
A strategy for cloning and mutagenesis of an infectious herpesvirus genome is described. The mouse cytomegalovirus genome was cloned and maintained as a 230 kb bacterial artificial chromosome (BAC) in E. coli. Transfection of the BAC plasmid into eukaryotic cells led to a productive virus infection. The feasibility to introduce targeted mutations into the BAC cloned virus genome was shown by mutation of the immediate-early 1 gene and generation of a mutant virus. Thus, the complete construction of a mutant herpesvirus genome can now be carried out in a controlled manner prior to the reconstitution of infectious progeny. The described approach should be generally applicable to the mutagenesis of genomes of other large DNA viruses.
Figures


References
-
- Britt W J, Alford C A. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. New York: Lippincott–Raven; 1996. pp. 2493–2523.
-
- Ho M, editor. Cytomegalovirus: Biology and Infection. New York: Plenum; 1991.
-
- Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Curr Top Microbiol Immunol. 1990;154:125–169. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

