Cloning-free PCR-based allele replacement methods
- PMID: 9414323
- PMCID: PMC310678
- DOI: 10.1101/gr.7.12.1174
Cloning-free PCR-based allele replacement methods
Abstract
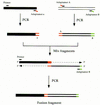
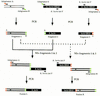
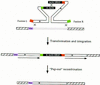
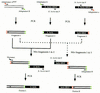
Efficient homologous recombination permits the directed introduction of specific mutations into the yeast genome. Here we describe a cloning-free, PCR-based allele replacement method that simplifies allele transfer between yeast strains. The desired allele from one strain is amplified by PCR, along with a selectable/counterselectable marker. After transformation, the resident allele in the target strain is replaced by creating a duplication of the new allele. Selection for direct repeat recombinants results in a single copy of the new allele in the target strain. Specifically, the desired allele is amplified by PCR with a pair of adaptamers, which are chimeric oligonucleotides that are used to amplify the allele and differentially tag its 5' and 3' ends. These tags allow the directed fusion to two different, but overlapping, regions of an appropriately tagged selectable/counterselectable marker after a second round of PCR amplification. Following cotransformation of the two fusion fragments into yeast, homologous recombination efficiently generates a duplication of the amplified allele flanking the intact selectable marker in the genome. After counterselection, only the desired allele is retained as a result of direct repeat recombination. A simple modification of this method allows the creation of de novo mutations in the genome.
Figures

Similar articles
-
Marker-fusion PCR for one-step mutagenesis of essential genes in yeast.Yeast. 2002 Jan 30;19(2):141-9. doi: 10.1002/yea.806. Yeast. 2002. PMID: 11788969
-
Polishing the craft of genetic diversity creation in directed evolution.Biotechnol Adv. 2013 Dec;31(8):1707-21. doi: 10.1016/j.biotechadv.2013.08.021. Epub 2013 Sep 6. Biotechnol Adv. 2013. PMID: 24012599 Review.
-
A new method for repeated "self-cloning" promoter replacement in Saccharomyces cerevisiae.Mol Biotechnol. 2011 Jul;48(3):218-27. doi: 10.1007/s12033-010-9362-6. Mol Biotechnol. 2011. PMID: 21170609
-
A two-step method for the introduction of single or multiple defined point mutations into the genome of Saccharomyces cerevisiae.Yeast. 2006 Aug;23(11):825-31. doi: 10.1002/yea.1397. Yeast. 2006. PMID: 16921548
-
Novel methods for cloning and engineering genes using the polymerase chain reaction.Curr Opin Biotechnol. 1995 Feb;6(1):30-6. doi: 10.1016/0958-1669(95)80006-9. Curr Opin Biotechnol. 1995. PMID: 7894080 Review.
Cited by
-
Immunity of the Saccharomyces cerevisiae SSY5 mRNA to nonsense-mediated mRNA decay.Front Mol Biosci. 2014 Dec 8;1:25. doi: 10.3389/fmolb.2014.00025. eCollection 2014. Front Mol Biosci. 2014. PMID: 25988166 Free PMC article.
-
Defects in the secretory pathway and high Ca2+ induce multiple P-bodies.Mol Biol Cell. 2010 Aug 1;21(15):2624-38. doi: 10.1091/mbc.e10-02-0099. Epub 2010 Jun 2. Mol Biol Cell. 2010. PMID: 20519435 Free PMC article.
-
Transient marker system for iterative gene targeting of a prototrophic fungus.Appl Environ Microbiol. 2007 Nov;73(22):7240-5. doi: 10.1128/AEM.01839-07. Epub 2007 Oct 5. Appl Environ Microbiol. 2007. PMID: 17921280 Free PMC article.
-
A conserved sequence extending motif III of the motor domain in the Snf2-family DNA translocase Rad54 is critical for ATPase activity.PLoS One. 2013 Dec 16;8(12):e82184. doi: 10.1371/journal.pone.0082184. eCollection 2013. PLoS One. 2013. PMID: 24358152 Free PMC article.
-
p53 Interacts with RNA polymerase II through its core domain and impairs Pol II processivity in vivo.PLoS One. 2011;6(8):e22183. doi: 10.1371/journal.pone.0022183. Epub 2011 Aug 4. PLoS One. 2011. PMID: 21829606 Free PMC article.
References
-
- Barnes WM. The fidelity of Taq polymerase catalyzing PCR is improved by an N-terminal deletion. Gene. 1992;112:29–35. - PubMed
-
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. - PubMed
-
- Erlich HA. PCR technology: Principles and applications for DNA amplification. New York, NY: Stockton Press; 1989.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
