The large subunit of basal transcription factor SNAPc is a Myb domain protein that interacts with Oct-1
- PMID: 9418884
- PMCID: PMC121507
- DOI: 10.1128/MCB.18.1.368
The large subunit of basal transcription factor SNAPc is a Myb domain protein that interacts with Oct-1
Abstract
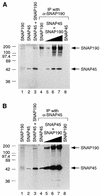
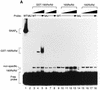
The human RNA polymerase II and III snRNA promoters have similar enhancers, the distal sequence elements (DSEs), and similar basal promoter elements, the proximal sequence elements (PSEs). The DSE, which contains an octamer motif, binds broadly expressed activator Oct-1. The PSE binds a multiprotein complex referred to as SNAPc or PTF. On DNAs containing both an octamer site and a PSE, Oct-1 and SNAPc bind cooperatively. SNAPc consists of at least four stably associated subunits, SNAP43, SNAP45, SNAP50, and SNAP190. None of the three small subunits, which have all been cloned, can bind to the PSE on their own. Here we report the isolation of cDNAs corresponding to the largest subunit of SNAPc, SNAP190. SNAP190 contains an unusual Myb DNA binding domain consisting of four complete repeats (Ra to Rd) and a half repeat (Rh). A truncated protein consisting of the last two SNAP190 Myb repeats, Rc and Rd, can bind to the PSE, suggesting that the SNAP190 Myb domain contributes to recognition of the PSE by the SNAP complex. SNAP190 is required for snRNA gene transcription by both RNA polymerases II and III and interacts with SNAP45. In addition, SNAP190 interacts with Oct-1. Together, these results suggest that the largest subunit of the SNAP complex is involved in direct recognition of the PSE and is a target for the Oct-1 activator. They also provide an example of a basal transcription factor containing a Myb DNA binding domain.
Figures







References
-
- Aasland R, Stewart A F, Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21:87–88. - PubMed
-
- Ford E, Hernandez N. Characterization of a trimeric complex containing Oct-1, SNAPc, and DNA. J Biol Chem. 1997;272:16048–16055. - PubMed
-
- Gstaiger M, Knoepfel L, Georgiev O, Schaffner W, Hovens C M. A B-cell coactivator of octamer-binding transcription factors. Nature. 1995;373:360–362. - PubMed
-
- Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
