Opposite transcriptional effects of cyclic AMP-responsive elements in confluent or p27KIP-overexpressing cells versus serum-starved or growing cells
- PMID: 9418888
- PMCID: PMC121511
- DOI: 10.1128/MCB.18.1.409
Opposite transcriptional effects of cyclic AMP-responsive elements in confluent or p27KIP-overexpressing cells versus serum-starved or growing cells
Abstract
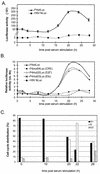
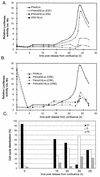
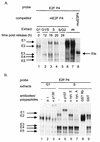
The minute virus of mice, an autonomous parvovirus, requires entry of host cells into the S phase of the cell cycle for its DNA to be amplified and its genes expressed. This work focuses on the P4 promoter of this parvovirus, which directs expression of the transcription unit encoding the parvoviral nonstructural polypeptides. These notably include protein NS1, necessary for the S-phase-dependent burst of parvoviral DNA amplification and gene expression. The activity of the P4 promoter is shown to be regulated in a cell cycle-dependent manner. At the G1/S-phase transition, the promoter is activated via a cis-acting DNA element which interacts with phase-specific complexes containing the cellular transcription factor E2F. It is inhibited, on the other hand, in cells arrested in G1 due to contact inhibition. This inhibitory effect is not observed in serum-starved cells. It is mediated in cis by cyclic AMP response elements (CREs). Unlike serum-starved cells, confluent cells accumulate the cyclin-dependent kinase inhibitor p27, suggesting that the switch from CRE-mediated activation to CRE-mediated repression involves the p27 protein. Accordingly, plasmid-driven overexpression of p27 causes down-modulation of promoter P4 in growing cells, depending on the presence of at least two functional CREs. No such effect is observed with two other cyclin-dependent kinase inhibitors, p16 and p21. Given the importance of P4-driven synthesis of protein NS1 in parvoviral DNA amplification and gene expression, the stringent S-phase dependency of promoter P4 is likely a major determinant of the absolute requirement of the minute virus of mice for host cell proliferation.
Figures

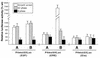



Similar articles
-
Activation of promoter P4 of the autonomous parvovirus minute virus of mice at early S phase is required for productive infection.J Virol. 1999 May;73(5):3877-85. doi: 10.1128/JVI.73.5.3877-3885.1999. J Virol. 1999. PMID: 10196282 Free PMC article.
-
Atypical nucleoprotein complexes mediate CRE-dependent regulation of the early promoter of minute virus of mice.J Gen Virol. 1999 Dec;80 ( Pt 12):3267-3272. doi: 10.1099/0022-1317-80-12-3267. J Gen Virol. 1999. PMID: 10567660
-
Assembly of cyclin D-dependent kinase and titration of p27Kip1 regulated by mitogen-activated protein kinase kinase (MEK1).Proc Natl Acad Sci U S A. 1998 Feb 3;95(3):1091-6. doi: 10.1073/pnas.95.3.1091. Proc Natl Acad Sci U S A. 1998. PMID: 9448290 Free PMC article.
-
E2F-1 accumulation bypasses a G1 arrest resulting from the inhibition of G1 cyclin-dependent kinase activity.Genes Dev. 1995 Dec 1;9(23):2873-87. doi: 10.1101/gad.9.23.2873. Genes Dev. 1995. PMID: 7498785
-
[Molecular mechanisms controlling the cell cycle: fundamental aspects and implications for oncology].Cancer Radiother. 2001 Apr;5(2):109-29. doi: 10.1016/s1278-3218(01)00087-7. Cancer Radiother. 2001. PMID: 11355576 Review. French.
Cited by
-
CREB/ATF-dependent repression of cyclin a by human T-cell leukemia virus type 1 Tax protein.J Virol. 2001 Mar;75(5):2161-73. doi: 10.1128/JVI.75.5.2161-2173.2001. J Virol. 2001. PMID: 11160720 Free PMC article.
-
Through its nonstructural protein NS1, parvovirus H-1 induces apoptosis via accumulation of reactive oxygen species.J Virol. 2010 Jun;84(12):5909-22. doi: 10.1128/JVI.01797-09. Epub 2010 Apr 7. J Virol. 2010. PMID: 20375165 Free PMC article.
-
Two new members of the emerging KDWK family of combinatorial transcription modulators bind as a heterodimer to flexibly spaced PuCGPy half-sites.Mol Cell Biol. 1999 Nov;19(11):7741-50. doi: 10.1128/MCB.19.11.7741. Mol Cell Biol. 1999. PMID: 10523663 Free PMC article.
-
Expression profiling of human hepatoma cells reveals global repression of genes involved in cell proliferation, growth, and apoptosis upon infection with parvovirus H-1.J Virol. 2005 Feb;79(4):2274-86. doi: 10.1128/JVI.79.4.2274-2286.2005. J Virol. 2005. PMID: 15681429 Free PMC article.
-
The Mammalian Cell Cycle Regulates Parvovirus Nuclear Capsid Assembly.PLoS Pathog. 2015 Jun 11;11(6):e1004920. doi: 10.1371/journal.ppat.1004920. eCollection 2015 Jun. PLoS Pathog. 2015. PMID: 26067441 Free PMC article.
References
-
- Ausubel I, Frederick M. Introduction of DNA into mammalian cells. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1987. pp. 9.7.12–9.7.18.
-
- Beijersbergen R L, Bernards R. Cell cycle regulation by the retinoblastoma family of growth inhibitory proteins. Biochim Biophys Acta. 1996;1287:103–120. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
