MYC abrogates p53-mediated cell cycle arrest in N-(phosphonacetyl)-L-aspartate-treated cells, permitting CAD gene amplification
- PMID: 9418900
- PMCID: PMC121521
- DOI: 10.1128/MCB.18.1.536
MYC abrogates p53-mediated cell cycle arrest in N-(phosphonacetyl)-L-aspartate-treated cells, permitting CAD gene amplification
Abstract
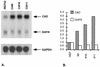
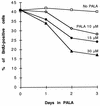
Genomic instability, including the ability to undergo gene amplification, is a hallmark of neoplastic cells. Similar to normal cells, "nonpermissive" REF52 cells do not develop resistance to N-(phosphonacetyl)-L-aspartate (PALA), an inhibitor of the synthesis of pyrimidine nucleotides, through amplification of cad, the target gene, but instead undergo protective, long-term, p53-dependent cell cycle arrest. Expression of exogenous MYC prevents this arrest and allows REF52 cells to proceed to mitosis when pyrimidine nucleotides are limiting. This results in DNA breaks, leading to cell death and, rarely, to cad gene amplification and PALA resistance. Pretreatment of REF52 cells with a low concentration of PALA, which slows DNA replication but does not trigger cell cycle arrest, followed by exposure to a high, selective concentration of PALA, promotes the formation of PALA-resistant cells in which the physically linked cad and endogenous N-myc genes are coamplified. The activated expression of endogenous N-myc in these pretreated PALA-resistant cells allows them to bypass the p53-mediated arrest that is characteristic of untreated REF52 cells. Our data demonstrate that two distinct events are required to form PALA-resistant REF52 cells: amplification of cad, whose product overcomes the action of the drug, and increased expression of N-myc, whose product overcomes the PALA-induced cell cycle block. These paired events occur at a detectable frequency only when the genes are physically linked, as cad and N-myc are. In untreated REF52 cells overexpressing N-MYC, the level of p53 is significantly elevated but there is no induction of p21waf1 expression or growth arrest. However, after DNA is damaged, the activated p53 executes rapid apoptosis in these REF52/N-myc cells instead of the long-term protective arrest seen in REF52 cells. The predominantly cytoplasmic localization of stabilized p53 in REF52/N-myc cells suggests that cytoplasmic retention may help to inactivate the growth-suppressing function of p53.
Figures





Similar articles
-
Late onset of CAD gene amplification in unamplified PALA resistant Chinese hamster mutants.Cancer Lett. 2000 Mar 31;150(2):119-27. doi: 10.1016/s0304-3835(99)00289-x. Cancer Lett. 2000. PMID: 10704733
-
p53-dependent growth arrest of REF52 cells containing newly amplified DNA.Proc Natl Acad Sci U S A. 1995 Apr 11;92(8):3224-8. doi: 10.1073/pnas.92.8.3224. Proc Natl Acad Sci U S A. 1995. PMID: 7724543 Free PMC article.
-
Multiple mechanisms of N-phosphonacetyl-L-aspartate resistance in human cell lines: carbamyl-P synthetase/aspartate transcarbamylase/dihydro-orotase gene amplification is frequent only when chromosome 2 is rearranged.Proc Natl Acad Sci U S A. 1997 Mar 4;94(5):1816-21. doi: 10.1073/pnas.94.5.1816. Proc Natl Acad Sci U S A. 1997. PMID: 9050862 Free PMC article.
-
New mechanisms of gene amplification in drug resistance (the episome model).Cancer Treat Res. 1991;57:1-11. doi: 10.1007/978-1-4615-3872-1_1. Cancer Treat Res. 1991. PMID: 1686710 Review. No abstract available.
-
CAD gene sequence and the domain structure of the mammalian multifunctional protein CAD.Biochem Soc Trans. 1993 Feb;21(1):186-91. doi: 10.1042/bst0210186. Biochem Soc Trans. 1993. PMID: 8095469 Review. No abstract available.
Cited by
-
Inactivation of p53 by human T-cell lymphotropic virus type 1 Tax requires activation of the NF-kappaB pathway and is dependent on p53 phosphorylation.Mol Cell Biol. 2000 May;20(10):3377-86. doi: 10.1128/MCB.20.10.3377-3386.2000. Mol Cell Biol. 2000. PMID: 10779327 Free PMC article.
-
Developmental context determines latency of MYC-induced tumorigenesis.PLoS Biol. 2004 Nov;2(11):e332. doi: 10.1371/journal.pbio.0020332. Epub 2004 Sep 28. PLoS Biol. 2004. PMID: 15455033 Free PMC article.
-
Length of time in tissue culture can affect the selected glyphosate resistance mechanism.Planta. 2004 Feb;218(4):589-98. doi: 10.1007/s00425-003-1130-4. Epub 2003 Oct 18. Planta. 2004. PMID: 14566562
-
A p53-dependent S-phase checkpoint helps to protect cells from DNA damage in response to starvation for pyrimidine nucleotides.Proc Natl Acad Sci U S A. 1998 Dec 8;95(25):14775-80. doi: 10.1073/pnas.95.25.14775. Proc Natl Acad Sci U S A. 1998. PMID: 9843965 Free PMC article.
-
Mitochondrial alteration in malignantly transformed human small airway epithelial cells induced by α-particles.Int J Cancer. 2013 Jan 1;132(1):19-28. doi: 10.1002/ijc.27656. Epub 2012 Jul 3. Int J Cancer. 2013. PMID: 22644783 Free PMC article.
References
-
- Agarwal, M. L., O. B. Chernova, A. Agarwal, W. R. Taylor, Y. K. Sharma, and G. R. Stark. Unpublished data.
-
- Alitalo K, Schwab M. Oncogene amplification in tumor cells. Adv Cancer Res. 1986;47:235–281. - PubMed
-
- Amati B, Land H. Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation and death. Curr Opin Genet Dev. 1994;4:102–108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
