Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin
- PMID: 9419339
- PMCID: PMC18146
- DOI: 10.1073/pnas.95.1.120
Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin
Abstract
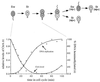
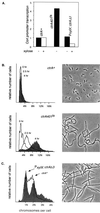
Caulobacter crescentus divides asymmetrically generating two distinct cell types at each cell division: a stalked cell competent for DNA replication, and a swarmer cell that is unable to initiate DNA replication until it differentiates into a stalked cell later in the cell cycle. The CtrA protein, a member of the response regulator family of the two-component signal transduction system, controls multiple cell cycle processes in Caulobacter and is present in swarmer cells but absent from stalked cells. We report that CtrA binds five sites within the chromosome replication origin in vitro. These sites overlap an essential DnaA box and a promoter in the origin that is essential for replication initiation. Analysis of mutant alleles of ctrA and point mutations in one of the CtrA binding sites in the origin demonstrate that CtrA represses replication in vivo. CtrA-mediated repression at the origin thus restricts replication to the stalked cell type. Thus, the direct coupling of chromosome replication with the cell cycle is mediated by the ubiquitous two-component signaling proteins.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

