Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads
- PMID: 9419361
- PMCID: PMC18190
- DOI: 10.1073/pnas.95.1.247
Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads
Abstract
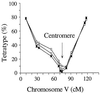
During meiosis, crossover events generate new allelic combinations, yet the abundance of these genetic exchanges in individual cells has not been measured previously on a genomic level. To perform a genome-wide analysis of recombination, we monitored the assortment of genetic markers in meiotic tetrads from Arabidopsis. By determining the number and distribution of crossovers in individual meiotic cells, we demonstrated (i) surprisingly precise regulation of crossover number in each meiosis, (ii) considerably reduced recombination along chromosomes carrying ribosomal DNA arrays, and (iii) an inversely proportional relationship between recombination frequencies and chromosome size. This use of tetrad analysis also achieved precise mapping of all five Arabidopsis centromeres, localizing centromere functions in the intact chromosomes of a higher eukaryote.
Figures



References
-
- Mather K, Beale G H. J Genetics. 1942;43:1–30.
-
- Petes T D, Malone R E, Symington L S. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Broach J R, Jones E S, Pringle J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1991. pp. 407–521.
-
- Carbon J, Clarke L. New Biol. 1990;2:10–19. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

