Large-scale temporal gene expression mapping of central nervous system development
- PMID: 9419376
- PMCID: PMC18216
- DOI: 10.1073/pnas.95.1.334
Large-scale temporal gene expression mapping of central nervous system development
Abstract
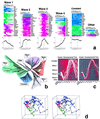
We used reverse transcription-coupled PCR to produce a high-resolution temporal map of fluctuations in mRNA expression of 112 genes during rat central nervous system development, focusing on the cervical spinal cord. The data provide a temporal gene expression "fingerprint" of spinal cord development based on major families of inter- and intracellular signaling genes. By using distance matrices for the pair-wise comparison of these 112 temporal gene expression patterns as the basis for a cluster analysis, we found five basic "waves" of expression that characterize distinct phases of development. The results suggest functional relationships among the genes fluctuating in parallel. We found that genes belonging to distinct functional classes and gene families clearly map to particular expression profiles. The concepts and data analysis discussed herein may be useful in objectively identifying coherent patterns and sequences of events in the complex genetic signaling network of development. Functional genomics approaches such as this may have applications in the elucidation of complex developmental and degenerative disorders.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

