Melanopsin: An opsin in melanophores, brain, and eye
- PMID: 9419377
- PMCID: PMC18217
- DOI: 10.1073/pnas.95.1.340
Melanopsin: An opsin in melanophores, brain, and eye
Abstract
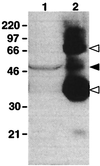
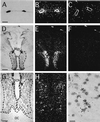
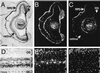
We have identified an opsin, melanopsin, in photosensitive dermal melanophores of Xenopus laevis. Its deduced amino acid sequence shares greatest homology with cephalopod opsins. The predicted secondary structure of melanopsin indicates the presence of a long cytoplasmic tail with multiple putative phosphorylation sites, suggesting that this opsin's function may be finely regulated. Melanopsin mRNA is expressed in hypothalamic sites thought to contain deep brain photoreceptors and in the iris, a structure known to be directly photosensitive in amphibians. Melanopsin message is also localized in retinal cells residing in the outermost lamina of the inner nuclear layer where horizontal cells are typically found. Its expression in retinal and nonretinal tissues suggests a role in vision and nonvisual photoreceptive tasks, such as photic control of skin pigmentation, pupillary aperture, and circadian and photoperiodic physiology.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

