Sequences within the herpesvirus-conserved pac1 and pac2 motifs are required for cleavage and packaging of the murine cytomegalovirus genome
- PMID: 9420199
- PMCID: PMC109348
- DOI: 10.1128/JVI.72.1.48-56.1998
Sequences within the herpesvirus-conserved pac1 and pac2 motifs are required for cleavage and packaging of the murine cytomegalovirus genome
Abstract
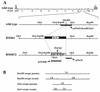
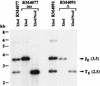
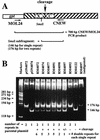
The DNA sequence motifs pac1 [an A-rich region flanked by poly(C) runs] and pac2 (CGCGGCG near an A-rich region) are conserved near herpesvirus genomic termini and are believed to mediate cleavage of genomes from replicative concatemers. To determine their importance in the cleavage process, we constructed a number of recombinant murine cytomegaloviruses with a second cleavage site inserted at an ectopic location within the viral genome. Cleavage at a wild-type ectopic site occurred as frequently as at the natural cleavage site, whereas mutation of this ectopic site revealed that some of the conserved motifs of pac1 and pac2 were essential for cleavage whereas others were not. Within pac1, the left poly(C) region was very important for cleavage and packaging but the A-rich region was not. Within pac2, the A-rich region and adjacent sequences were essential for cleavage and packaging and the CGCGGCG region contributed to, but was not strictly essential for, efficient cleavage and packaging. A second A-rich region was not important at all. Furthermore, mutations that prevented cleavage also blocked duplication and deletion of the murine cytomegalovirus 30-bp terminal repeat at the ectopic site, suggesting that repeat duplication and deletion are consequences of cleavage. Given that the processes of genome cleavage and packaging appear to be highly conserved among herpesviruses, these findings should be relevant to other members of this family.
Figures




Similar articles
-
The ends on herpesvirus DNA replicative concatemers contain pac2 cis cleavage/packaging elements and their formation is controlled by terminal cis sequences.J Virol. 2000 Feb;74(3):1587-92. doi: 10.1128/jvi.74.3.1587-1592.2000. J Virol. 2000. PMID: 10627574 Free PMC article.
-
Functional identification and analysis of cis-acting sequences which mediate genome cleavage and packaging in human herpesvirus 6.J Virol. 1998 Jan;72(1):320-9. doi: 10.1128/JVI.72.1.320-329.1998. J Virol. 1998. PMID: 9420230 Free PMC article.
-
A 128-base-pair sequence containing the pac1 and a presumed cryptic pac2 sequence includes cis elements sufficient to mediate efficient genome maturation of human cytomegalovirus.J Virol. 2011 May;85(9):4432-9. doi: 10.1128/JVI.02307-10. Epub 2011 Feb 23. J Virol. 2011. PMID: 21345955 Free PMC article.
-
Human cytomegalovirus terminase as a target for antiviral chemotherapy.Rev Med Virol. 2002 Mar-Apr;12(2):115-27. doi: 10.1002/rmv.344. Rev Med Virol. 2002. PMID: 11921307 Review.
-
Nonrandom clusters of palindromes in herpesvirus genomes.J Comput Biol. 2005 Apr;12(3):331-54. doi: 10.1089/cmb.2005.12.331. J Comput Biol. 2005. PMID: 15857246 Free PMC article. Review.
Cited by
-
The ends on herpesvirus DNA replicative concatemers contain pac2 cis cleavage/packaging elements and their formation is controlled by terminal cis sequences.J Virol. 2000 Feb;74(3):1587-92. doi: 10.1128/jvi.74.3.1587-1592.2000. J Virol. 2000. PMID: 10627574 Free PMC article.
-
Structural variability of the herpes simplex virus 1 genome in vitro and in vivo.J Virol. 2012 Aug;86(16):8592-601. doi: 10.1128/JVI.00223-12. Epub 2012 Jun 6. J Virol. 2012. PMID: 22674981 Free PMC article.
-
Effects of mutations within the herpes simplex virus type 1 DNA encapsidation signal on packaging efficiency.J Virol. 2001 Oct;75(19):8977-86. doi: 10.1128/JVI.75.19.8977-8986.2001. J Virol. 2001. PMID: 11533161 Free PMC article.
-
The impact of genome length on replication and genome stability of the herpesvirus guinea pig cytomegalovirus.Virology. 2009 Mar 30;386(1):132-8. doi: 10.1016/j.virol.2008.12.030. Epub 2009 Jan 26. Virology. 2009. PMID: 19174305 Free PMC article.
-
Analysis of herpes simplex virus type 1 DNA packaging signal mutations in the context of the viral genome.J Virol. 2010 Jan;84(1):321-9. doi: 10.1128/JVI.01489-09. J Virol. 2010. PMID: 19864384 Free PMC article.
References
-
- Addison C, Rixon F J, Palfreyman J W, O’Hara M, Preston V G. Characterisation of a herpes simplex virus type 1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology. 1984;138:246–259. - PubMed
-
- Addison C, Rixon F J, Preston V G. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J Gen Virol. 1990;71:2377–2384. - PubMed
-
- Al-Kobaisi M F, Rixon F J, McDougall I, Preston V G. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology. 1991;180:380–388. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

