Genetic analysis of the diversity and origin of hantaviruses in Peromyscus leucopus mice in North America
- PMID: 9420200
- PMCID: PMC109349
- DOI: 10.1128/JVI.72.1.57-64.1998
Genetic analysis of the diversity and origin of hantaviruses in Peromyscus leucopus mice in North America
Abstract
Nucleotide sequences were determined for the complete M genome segments of two distinct hantavirus genetic lineages which were detected in hantavirus antibody- and PCR-positive white-footed mice (Peromyscus leucopus) from Indiana and Oklahoma. Phylogenetic analyses indicated that although divergent from each other, the virus lineages in Indiana and Oklahoma were monophyletic and formed a newly identified unique ancestral branch within the clade of Sin Nombre-like viruses found in Peromyscus mice. Interestingly, P. leucopus-borne New York virus was found to be most closely related to the P. maniculatus-borne viruses, Sin Nombre and Monongahela, and monophyletic with Monongahela virus. In parallel, intraspecific phylogenetic relationships of P. leucopus were also determined, based on the amplification, sequencing, and analysis of the DNA fragment representing the replication control region of the rodent mitochondrial genome. P. leucopus mitochondrial DNA haplotypes were found to form four separate genetic clades, referred to here as Eastern, Central, Northwestern, and Southwestern groups. The distinct Indiana and Oklahoma virus lineages were detected in P. leucopus of the Eastern and Southwestern mitochondrial DNA haplotypes, respectively. Taken together, our current data suggests that both cospeciation of Peromyscus-borne hantaviruses with their specific rodent hosts and biogeographic factors (such as allopatric migrations, geographic separation, and isolation) have played important roles in establishment of the current genetic diversity and geographic distribution of Sin Nombre-like hantaviruses. In particular, the unusual position of New York virus on the virus phylogenetic tree is most consistent with an historically recent host-switching event.
Figures


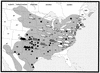
References
-
- Anderson S, Bankier A T, Barrell B G, de Bruijn M H L, Coulson A R, Drouin J, Eperon I C, Nierlich D P, Roe B A, Sanger F, Schreier P H, Smith A J H, Staden R, Young I G. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
-
- Antic D, Yong Kang C, Spik K, Schmaljohn C S, Vapalahti O, Vaheri A. Comparison of the deduced gene products of the L, M and S genome segments of hantaviruses. Virus Res. 1992;24:35–46. - PubMed
-
- Arikawa J, Lapenotiere H F, Iacono-Connors L, Wang M, Schmaljohn C S. Coding properties of the S and M genome segments of Sapporo rat virus: comparison to other causative agents of hemorrhagic fever with renal syndrome. Virology. 1990;176:114–125. - PubMed
-
- Baker R H. Habitats and distribution. In: King J A, editor. Biology of Peromyscus (Rodentia). Special publication. Lawrence, Kans: American Society of Mammalogists; 1968. pp. 98–126.
-
- Baker R J, Robbins L W, Stangl F B, Jr, Birney E C. Chromosomal evidence for a major subdivision in Peromyscus leucopus. J Mammal. 1983;64:356–359.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

